7MCU
 
 | | Crystal structure of Staphylococcus aureus Cystathionine gamma lyase, Holoenzyme with bound NL2 | | Descriptor: | 5-[(6-bromo-1H-indol-1-yl)methyl]-2-methylfuran-3-carboxylic acid, Bifunctional cystathionine gamma-lyase/homocysteine desulfhydrase, CITRATE ANION, ... | | Authors: | Nuthanakanti, A, Serganov, A, Kaushik, A. | | Deposit date: | 2021-04-02 | | Release date: | 2021-06-23 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Inhibitors of bacterial H 2 S biogenesis targeting antibiotic resistance and tolerance.
Science, 372, 2021
|
|
7MCT
 
 | | Crystal structure of Staphylococcus aureus Cystathionine gamma lyase, Holoenzyme with bound NL1 | | Descriptor: | Bifunctional cystathionine gamma-lyase/homocysteine desulfhydrase, GLYCEROL, N-[(6-bromo-1H-indol-1-yl)acetyl]glycine, ... | | Authors: | Nuthanakanti, A, Serganov, A, Kaushik, A. | | Deposit date: | 2021-04-02 | | Release date: | 2021-06-23 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Inhibitors of bacterial H 2 S biogenesis targeting antibiotic resistance and tolerance.
Science, 372, 2021
|
|
7MD8
 
 | | Crystal structure of Staphylococcus aureus cystathionine gamma lyase holoenzyme Y103N mutant co-crystallized with NL2 | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, Bifunctional cystathionine gamma-lyase/homocysteine desulfhydrase, GLYCEROL, ... | | Authors: | Nuthanakanti, A, Serganov, A, Kaushik, A. | | Deposit date: | 2021-04-03 | | Release date: | 2021-06-23 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.06 Å) | | Cite: | Inhibitors of bacterial H 2 S biogenesis targeting antibiotic resistance and tolerance.
Science, 372, 2021
|
|
7MDA
 
 | | Crystal structure of Staphylococcus aureus cystathionine gamma lyase holoenzyme Y103A mutant co-crystallized with NL1 | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, Bifunctional cystathionine gamma-lyase/homocysteine desulfhydrase, N-[(6-bromo-1H-indol-1-yl)acetyl]glycine, ... | | Authors: | Nuthanakanti, A, Serganov, A, Kaushik, A. | | Deposit date: | 2021-04-03 | | Release date: | 2021-06-23 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.24 Å) | | Cite: | Inhibitors of bacterial H 2 S biogenesis targeting antibiotic resistance and tolerance.
Science, 372, 2021
|
|
9C87
 
 | |
9C88
 
 | |
7WX7
 
 | | complex of a legionella acetyltransferase VipF and COA/ACO | | Descriptor: | ACETYL COENZYME *A, COENZYME A, N-acetyltransferase | | Authors: | Chen, T.T, Lin, Y.L, Zhang, S.J, Han, A.D. | | Deposit date: | 2022-02-14 | | Release date: | 2023-02-22 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.781 Å) | | Cite: | Structural basis for the acetylation mechanism of the Legionella effector VipF.
Acta Crystallogr D Struct Biol, 78, 2022
|
|
7WX5
 
 | | a Legionella acetyltransferase effector VipF | | Descriptor: | ACETYL COENZYME *A, N-acetyltransferase | | Authors: | Chen, T.T, Lin, Y.L, Zhang, S.J, Han, A.D. | | Deposit date: | 2022-02-14 | | Release date: | 2023-02-22 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.392 Å) | | Cite: | Structural basis for the acetylation mechanism of the Legionella effector VipF.
Acta Crystallogr D Struct Biol, 78, 2022
|
|
2A07
 
 | | Crystal Structure of Foxp2 bound Specifically to DNA. | | Descriptor: | 5'-D(*AP*AP*CP*TP*AP*TP*GP*AP*AP*AP*CP*AP*AP*AP*TP*TP*TP*TP*CP*CP*T)-3', 5'-D(*TP*TP*AP*GP*GP*AP*AP*AP*AP*TP*TP*TP*GP*TP*TP*TP*CP*AP*TP*AP*G)-3', Forkhead box protein P2, ... | | Authors: | Stroud, J.C, Wu, Y, Bates, D.L, Han, A, Nowick, K, Paabo, S, Tong, H, Chen, L. | | Deposit date: | 2005-06-16 | | Release date: | 2006-01-31 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structure of the Forkhead Domain of FOXP2 Bound to DNA.
Structure, 14, 2006
|
|
5K18
 
 | | The NatB Acetyltransferase Complex Bound To bisubstrate inhibitor | | Descriptor: | Bisubstrate inhibitor, COENZYME A, N-terminal acetyltransferase B complex subunit NAT3, ... | | Authors: | Hong, H, Cai, Y, Zhang, S, Han, A. | | Deposit date: | 2016-05-17 | | Release date: | 2017-04-19 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.73 Å) | | Cite: | Molecular Basis of Substrate Specific Acetylation by N-Terminal Acetyltransferase NatB
Structure, 25, 2017
|
|
5K04
 
 | | The NatB Acetyltransferase Complex Bound To CoA and MES | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, COENZYME A, N-terminal acetyltransferase B complex subunit NAT3, ... | | Authors: | Hong, H, Cai, Y, Zhang, S, Han, A. | | Deposit date: | 2016-05-17 | | Release date: | 2017-04-19 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Molecular Basis of Substrate Specific Acetylation by N-Terminal Acetyltransferase NatB
Structure, 25, 2017
|
|
5TUD
 
 | | Structural Insights into the Extracellular Recognition of the Human Serotonin 2B Receptor by an Antibody | | Descriptor: | 5-hydroxytryptamine receptor 2B,Soluble cytochrome b562 chimera, Anti-5-HT2B Fab heavy chain, Anti-5-HT2B Fab light chain, ... | | Authors: | Ishchenko, A, Wacker, D, Kapoor, M, Zhang, A, Han, G.W, Basu, S, Boutet, S, James, D, Wang, D, Weierstall, U, Liu, W, Katritch, V, Stevens, R.C, Cherezov, V. | | Deposit date: | 2016-11-05 | | Release date: | 2017-07-26 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structural insights into the extracellular recognition of the human serotonin 2B receptor by an antibody.
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
4GQB
 
 | | Crystal Structure of the human PRMT5:MEP50 Complex | | Descriptor: | (2S,5S,6E)-2,5-diamino-6-[(3S,4R,5R)-5-(6-amino-9H-purin-9-yl)-3,4-dihydroxydihydrofuran-2(3H)-ylidene]hexanoic acid, Histone H4 peptide, Methylosome protein 50, ... | | Authors: | Antonysamy, S, Bonday, Z, Campbell, R, Doyle, B, Druzina, Z, Gheyi, T, Han, B, Jungheim, L.N, Qian, Y, Rauch, C, Russell, M, Sauder, J.M, Wasserman, S.R, Weichert, K, Willard, F.S, Zhang, A, Emtage, S. | | Deposit date: | 2012-08-22 | | Release date: | 2012-10-17 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.06 Å) | | Cite: | Crystal structure of the human PRMT5:MEP50 complex.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
8H24
 
 | | Leucine-rich alpha-2-glycoprotein 1 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Leucine-rich alpha-2-glycoprotein, SULFATE ION | | Authors: | Won, S.Y, Park, B.S, Lee, D.S, Kim, H.M, Han, A, Yang, J. | | Deposit date: | 2022-10-04 | | Release date: | 2023-08-23 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Crystal structure of LRG1 and the functional significance of LRG1 glycan for LPHN2 activation.
Exp.Mol.Med., 55, 2023
|
|
7C0Q
 
 | | a Legionella pneumophila effector Lpg2505 | | Descriptor: | GLYCEROL, effector Lpg2505 | | Authors: | Chen, T.T, Han, A.D, Luo, Z.Q, McCloskey, A, Perri, K. | | Deposit date: | 2020-05-01 | | Release date: | 2021-02-24 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | The metaeffector MesI regulates the activity of the Legionella effector SidI through direct protein-protein interactions.
Microbes Infect., 23, 2021
|
|
7WX6
 
 | | A Legionella acetyltransferase VipF | | Descriptor: | CHLORAMPHENICOL, COENZYME A, N-acetyltransferase | | Authors: | Chen, T.T, Lin, Y.L, Chen, Z, Han, A.D. | | Deposit date: | 2022-02-14 | | Release date: | 2022-09-14 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.273 Å) | | Cite: | Structural basis for the acetylation mechanism of the Legionella effector VipF.
Acta Crystallogr D Struct Biol, 78, 2022
|
|
7E72
 
 | | Crystal structure of Tie2-agonistic antibody in complex with human Tie2 Fn2-3 | | Descriptor: | 1,2-ETHANEDIOL, Angiopoietin-1 receptor, the chimeric Fab fragment of 3H7 (heavy chain), ... | | Authors: | Kim, H.M, Jo, G.H, Hong, H.J, Han, A. | | Deposit date: | 2021-02-25 | | Release date: | 2021-11-10 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.094 Å) | | Cite: | Structural insights into the clustering and activation of Tie2 receptor mediated by Tie2 agonistic antibody.
Nat Commun, 12, 2021
|
|
4Q2Y
 
 | | Crystal structure of Arginyl-tRNA synthetase | | Descriptor: | Arginine--tRNA ligase, cytoplasmic | | Authors: | Kim, H.S, Jo, C.H, Cha, S.Y, Han, A.R, Hwang, K.Y. | | Deposit date: | 2014-04-10 | | Release date: | 2014-07-23 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.799 Å) | | Cite: | The crystal structure of arginyl-tRNA synthetase from Homo sapiens
Febs Lett., 588, 2014
|
|
4Q2T
 
 | | Crystal structure of Arginyl-tRNA synthetase complexed with L-arginine | | Descriptor: | ARGININE, Arginine--tRNA ligase, cytoplasmic, ... | | Authors: | Kim, H.S, Jo, C.H, Cha, S.Y, Han, A.R, Hwang, K.Y. | | Deposit date: | 2014-04-09 | | Release date: | 2014-07-23 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | The crystal structure of arginyl-tRNA synthetase from Homo sapiens
Febs Lett., 588, 2014
|
|
5ZA3
 
 | |
4Q2X
 
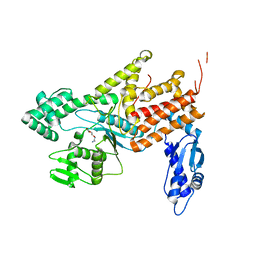 | | Crystal structure of Arginyl-tRNA synthetase complexed with L-canavanine | | Descriptor: | Arginine--tRNA ligase, cytoplasmic, L-CANAVANINE | | Authors: | Kim, H.S, Jo, C.H, Cha, S.Y, Han, A.R, Hwang, K.Y. | | Deposit date: | 2014-04-10 | | Release date: | 2014-07-23 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.798 Å) | | Cite: | The crystal structure of arginyl-tRNA synthetase from Homo sapiens
Febs Lett., 588, 2014
|
|
5DN8
 
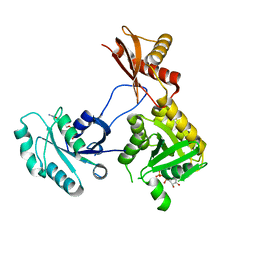 | | 1.76 Angstrom Crystal Structure of GTP-binding Protein Der from Coxiella burnetii in Complex with GDP. | | Descriptor: | GTPase Der, GUANOSINE-5'-DIPHOSPHATE | | Authors: | Minasov, G, Shuvalova, L, Han, A, Kim, H.-Y, Grimshaw, S, Kwon, K, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2015-09-09 | | Release date: | 2015-10-14 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.76 Å) | | Cite: | 1.76 Angstrom Crystal Structure of GTP-binding Protein Der from Coxiella burnetii in Complex with GDP.
To Be Published
|
|
3KOV
 
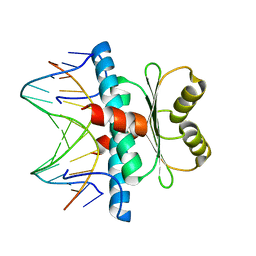 | | Structure of MEF2A bound to DNA reveals a completely folded MADS-box/MEF2 domain that recognizes DNA and recruits transcription co-factors | | Descriptor: | DNA (5'-D(*AP*AP*CP*TP*AP*TP*TP*TP*AP*TP*AP*AP*G)-3'), DNA (5'-D(*TP*CP*TP*TP*AP*TP*AP*AP*AP*TP*AP*GP*T)-3'), Myocyte-specific enhancer factor 2A | | Authors: | Wu, Y, Dey, R, Han, A, Jayathilaka, N, Philips, M, Ye, J, Chen, L. | | Deposit date: | 2009-11-14 | | Release date: | 2010-02-16 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structure of the MADS-box/MEF2 Domain of MEF2A Bound to DNA and Its Implication for Myocardin Recruitment.
J.Mol.Biol., 397, 2010
|
|
3MU6
 
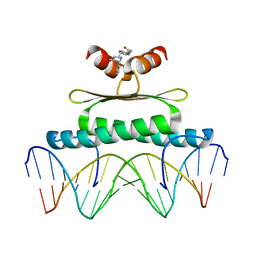 | | Inhibiting the Binding of Class IIa Histone Deacetylases to Myocyte Enhancer Factor-2 by Small Molecules | | Descriptor: | (3E)-N~8~-(2-aminophenyl)-N~1~-phenyloct-3-enediamide, DNA (5'-D(*AP*AP*AP*GP*CP*TP*AP*TP*TP*AP*TP*TP*AP*GP*CP*TP*T)-3'), DNA (5'-D(*TP*AP*AP*GP*CP*TP*AP*AP*TP*AP*AP*TP*AP*GP*CP*TP*T)-3'), ... | | Authors: | Jayathilaka, N, Han, A, Gaffney, K, Dey, R, He, J, Ye, J, Gao, T, Petasis, N.A, Chen, L. | | Deposit date: | 2010-05-01 | | Release date: | 2011-11-02 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.434 Å) | | Cite: | Inhibition of the function of class IIa HDACs by blocking their interaction with MEF2.
Nucleic Acids Res., 40, 2012
|
|
2AS5
 
 | | Structure of the DNA binding domains of NFAT and FOXP2 bound specifically to DNA. | | Descriptor: | 5'-D(AP*AP*CP*TP*AP*TP*GP*AP*AP*AP*CP*AP*AP*AP*TP*TP*TP*TP*CP*CP*TP*)-3', 5'-D(TP*TP*AP*GP*GP*AP*AP*AP*AP*TP*TP*TP*GP*TP*TP*TP*CP*AP*TP*AP*GP*)-3', Forkhead box protein P2, ... | | Authors: | Wu, Y, Stroud, J.C, Borde, M, Bates, D.L, Guo, L, Han, A, Rao, A, Chen, L. | | Deposit date: | 2005-08-22 | | Release date: | 2006-08-08 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | FOXP3 Controls Regulatory T Cell Function through Cooperation with NFAT.
Cell(Cambridge,Mass.), 126, 2006
|
|
