7E5M
 
 | | crystal structure of trans assembled human TROP-2 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Tumor-associated calcium signal transducer 2 | | Authors: | Sun, M, Zhang, H, Chai, Y, Qi, J, Gao, G.F, Tan, S. | | Deposit date: | 2021-02-19 | | Release date: | 2021-12-15 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Structural insights into the cis and trans assembly of human trophoblast cell surface antigen 2.
Iscience, 24, 2021
|
|
7E5N
 
 | | crystal structure of cis assembled TROP-2 | | Descriptor: | Tumor-associated calcium signal transducer 2 | | Authors: | Sun, M, Zhang, H, Chai, Y, Qi, J, Gao, G.F, Tan, S. | | Deposit date: | 2021-02-19 | | Release date: | 2021-12-15 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Structural insights into the cis and trans assembly of human trophoblast cell surface antigen 2.
Iscience, 24, 2021
|
|
3A8Q
 
 | | Low-resolution crystal structure of the Tiam2 PHCCEx domain | | Descriptor: | T-lymphoma invasion and metastasis-inducing protein 2 | | Authors: | Terawaki, S, Kitano, K, Mori, T, Zhai, Y, Higuchi, Y, Itoh, N, Watanabe, T, Kaibuchi, K, Hakoshima, T. | | Deposit date: | 2009-10-07 | | Release date: | 2009-11-24 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | The PHCCEx domain of Tiam1/2 is a novel protein- and membrane-binding module
Embo J., 29, 2010
|
|
2GA6
 
 | | The crystal structure of SARS nsp10 without zinc ion as additive | | Descriptor: | ZINC ION, orf1a polyprotein | | Authors: | Su, D, Lou, Z, Sun, F, Zhai, Y, Yang, H, Rao, Z. | | Deposit date: | 2006-03-08 | | Release date: | 2006-08-15 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Dodecamer Structure of Severe Acute Respiratory Syndrome Coronavirus Nonstructural Protein nsp10
J.Virol., 80, 2006
|
|
3A2A
 
 | | The structure of the carboxyl-terminal domain of the human voltage-gated proton channel Hv1 | | Descriptor: | CHLORIDE ION, Voltage-gated hydrogen channel 1 | | Authors: | Li, S.J, Unno, H, Zhou, Q, Zhao, Q, Zhai, Y, Sun, F. | | Deposit date: | 2009-05-08 | | Release date: | 2010-02-09 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The role and structure of the carboxyl-terminal domain of the human voltage-gated proton channel Hv1.
J.Biol.Chem., 285, 2010
|
|
3SWF
 
 | | CNGA1 621-690 containing CLZ domain | | Descriptor: | ZINC ION, cGMP-gated cation channel alpha-1 | | Authors: | Shuart, N.G, Haitin, Y, Camp, S.S, Black, K.D, Zagotta, W.N. | | Deposit date: | 2011-07-13 | | Release date: | 2011-09-14 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.14 Å) | | Cite: | Molecular mechanism for 3:1 subunit stoichiometry of rod cyclic nucleotide-gated ion channels.
Nat Commun, 2, 2011
|
|
3SWY
 
 | | CNGA3 626-672 containing CLZ domain | | Descriptor: | Cyclic nucleotide-gated cation channel alpha-3 | | Authors: | Shuart, N.G, Haitin, Y, Camp, S.S, Black, K.D, Zagotta, W.N. | | Deposit date: | 2011-07-14 | | Release date: | 2011-09-14 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Molecular mechanism for 3:1 subunit stoichiometry of rod cyclic nucleotide-gated ion channels.
Nat Commun, 2, 2011
|
|
1ZP0
 
 | | Crystal Structure of Mitochondrial Respiratory Complex II bound with 3-nitropropionate and 2-thenoyltrifluoroacetone | | Descriptor: | 3-NITROPROPANOIC ACID, 4,4,4-TRIFLUORO-1-THIEN-2-YLBUTANE-1,3-DIONE, FAD-binding protein, ... | | Authors: | Sun, F, Huo, X, Zhai, Y, Wang, A, Xu, J, Su, D, Bartlam, M, Rao, Z. | | Deposit date: | 2005-05-16 | | Release date: | 2005-07-12 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Crystal Structure of Mitochondrial Respiratory Membrane Protein Complex II
Cell(Cambridge,Mass.), 121, 2005
|
|
1ZOY
 
 | | Crystal Structure of Mitochondrial Respiratory Complex II from porcine heart at 2.4 Angstroms | | Descriptor: | FAD-binding protein, FE2/S2 (INORGANIC) CLUSTER, FE3-S4 CLUSTER, ... | | Authors: | Sun, F, Huo, X, Zhai, Y, Wang, A, Xu, J, Su, D, Bartlam, M, Rao, Z. | | Deposit date: | 2005-05-15 | | Release date: | 2005-07-12 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal Structure of Mitochondrial Respiratory Membrane Protein Complex II
Cell(Cambridge,Mass.), 121, 2005
|
|
7XNF
 
 | | Structure of SARS-CoV-2 antibody P2C-1F11 with GX/P2V/2017 RBD | | Descriptor: | P2C-1F11 Heavy Chain, P2C-1F11 Lambda chain, Spike protein S1 | | Authors: | Jia, Y.F, Chai, Y, Wang, Q.H, Gao, G.F. | | Deposit date: | 2022-04-28 | | Release date: | 2023-01-11 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.79 Å) | | Cite: | Cross-reaction of current available SARS-CoV-2 MAbs against the pangolin-origin coronavirus GX/P2V/2017.
Cell Rep, 41, 2022
|
|
7W1Y
 
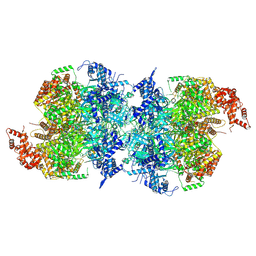 | | Human MCM double hexamer bound to natural DNA duplex (polyAT/polyTA) | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, DNA (49-MER), ... | | Authors: | Li, J, Dong, J, Dang, S, Zhai, Y. | | Deposit date: | 2021-11-21 | | Release date: | 2023-02-08 | | Last modified: | 2024-06-26 | | Method: | ELECTRON MICROSCOPY (2.59 Å) | | Cite: | The human pre-replication complex is an open complex.
Cell, 186, 2023
|
|
7XSW
 
 | | Structure of SARS-CoV-2 antibody S309 with GX/P2V/2017 RBD | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, S309 Heavy Chain, S309 Lambda Chain, ... | | Authors: | Jia, Y.F, Chai, Y, Wang, Q.H, Gao, G.F. | | Deposit date: | 2022-05-15 | | Release date: | 2023-01-11 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Cross-reaction of current available SARS-CoV-2 MAbs against the pangolin-origin coronavirus GX/P2V/2017.
Cell Rep, 41, 2022
|
|
7YE7
 
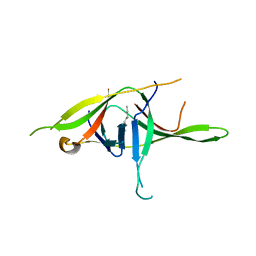 | | Crystal structure of SARS-CoV-2 soluble dimeric ORF9b | | Descriptor: | N-OCTANE, ORF9b protein, nonane | | Authors: | Jin, X, Chai, Y, Qi, J, Song, H, Gao, G.F. | | Deposit date: | 2022-07-05 | | Release date: | 2022-10-19 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.95 Å) | | Cite: | Structural characterization of SARS-CoV-2 dimeric ORF9b reveals potential fold-switching trigger mechanism.
Sci China Life Sci, 66, 2023
|
|
7YE8
 
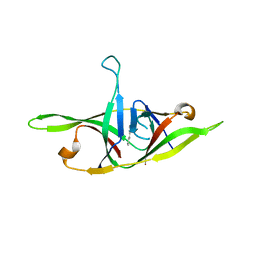 | | Crystal structure of SARS-CoV-2 refolded dimeric ORF9b | | Descriptor: | N-OCTANE, ORF9b protein | | Authors: | Jin, X, Chai, Y, Qi, J, Song, H, Gao, G.F. | | Deposit date: | 2022-07-05 | | Release date: | 2022-10-19 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (3.01 Å) | | Cite: | Structural characterization of SARS-CoV-2 dimeric ORF9b reveals potential fold-switching trigger mechanism.
Sci China Life Sci, 66, 2023
|
|
7V3U
 
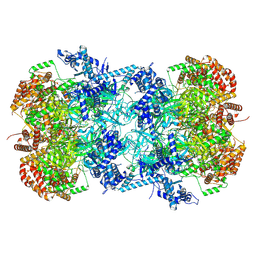 | | Cryo-EM structure of MCM double hexamer with structured Mcm4-NSD | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, DNA replication licensing factor MCM2, DNA replication licensing factor MCM3, ... | | Authors: | Cheng, J, Li, N, Huo, Y, Dang, S, Tye, B, Gao, N, Zhai, Y. | | Deposit date: | 2021-08-11 | | Release date: | 2022-04-13 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Structural Insight into the MCM double hexamer activation by Dbf4-Cdc7 kinase.
Nat Commun, 13, 2022
|
|
7V3V
 
 | | Cryo-EM structure of MCM double hexamer bound with DDK in State I | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Cell division control protein 7, DDK kinase regulatory subunit DBF4, ... | | Authors: | Cheng, J, Li, N, Huo, Y, Dang, S, Tye, B, Gao, N, Zhai, Y. | | Deposit date: | 2021-08-11 | | Release date: | 2022-04-13 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Structural Insight into the MCM double hexamer activation by Dbf4-Cdc7 kinase.
Nat Commun, 13, 2022
|
|
7W8G
 
 | | Cryo-EM structure of MCM double hexamer | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, DNA replication licensing factor MCM2, DNA replication licensing factor MCM3, ... | | Authors: | Cheng, J, Li, N, Tye, B, Zhai, Y, Gao, N. | | Deposit date: | 2021-12-07 | | Release date: | 2022-04-13 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (2.52 Å) | | Cite: | Structural Insight into the MCM double hexamer activation by Dbf4-Cdc7 kinase.
Nat Commun, 13, 2022
|
|
7Y3N
 
 | | Crystal structure of SARS-CoV receptor binding domain in complex with human antibody BIOLS56 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, Heavy chain of BIOLS56, ... | | Authors: | Rao, X, Chai, Y, Wu, Y, Gao, F. | | Deposit date: | 2022-06-11 | | Release date: | 2023-12-27 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.97 Å) | | Cite: | Defining a de novo non-RBM antibody as RBD-8 and its synergistic rescue of immune-evaded antibodies to neutralize Omicron SARS-CoV-2.
Proc.Natl.Acad.Sci.USA, 120, 2023
|
|
1R1Y
 
 | | Crystal structure of deoxy-human hemoglobin Bassett at 1.8 angstrom | | Descriptor: | Hemoglobin alpha chain, Hemoglobin beta chain, PROTOPORPHYRIN IX CONTAINING FE, ... | | Authors: | Abdulmalik, O, Safo, M.K, Lerner, N.B, Ochotorena, J, Dhaikin, Y, Abraham, D.J, Asakura, T. | | Deposit date: | 2003-09-25 | | Release date: | 2003-10-07 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Characterization of hemoglobin bassett (alpha94Asp-->Ala), a variant with very low oxygen affinity
Am.J.Hematol., 77, 2004
|
|
1R1X
 
 | | Crystal structure of oxy-human hemoglobin Bassett at 2.15 angstrom | | Descriptor: | CARBON MONOXIDE, Hemoglobin alpha chain, Hemoglobin beta chain, ... | | Authors: | Abdulmalik, O, Safo, M.K, Lerner, N.B, Ochotorena, J, Dhaikin, Y, Abraham, D.J, Asakura, T. | | Deposit date: | 2003-09-25 | | Release date: | 2003-10-07 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Characterization of hemoglobin bassett (alpha94Asp-->Ala), a variant with very low oxygen affinity
Am.J.Hematol., 77, 2004
|
|
8YAJ
 
 | | ATAT-2 bound MEC-12/MEC-7 microtubule without acetyl-CoA | | Descriptor: | Alpha-tubulin N-acetyltransferase 2, GUANOSINE-5'-TRIPHOSPHATE, PHOSPHOMETHYLPHOSPHONIC ACID GUANYLATE ESTER, ... | | Authors: | Lam, W.H, Yu, D, Zhai, Y, Ti, S. | | Deposit date: | 2024-02-09 | | Release date: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Tubulin acetyltransferases access and modify the microtubule luminal K40 residue through anchors in taxane-binding pockets.
Nat.Struct.Mol.Biol., 2024
|
|
8YAR
 
 | | ATAT-2 bound K40R MEC-12/MEC-7 microtubule | | Descriptor: | ACETYL COENZYME *A, Alpha-tubulin N-acetyltransferase 2, GUANOSINE-5'-TRIPHOSPHATE, ... | | Authors: | Lam, W.H, Yu, D, Zhai, Y, Ti, S. | | Deposit date: | 2024-02-09 | | Release date: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Tubulin acetyltransferases access and modify the microtubule luminal K40 residue through anchors in taxane-binding pockets.
Nat.Struct.Mol.Biol., 2024
|
|
8Y9F
 
 | | ATAT-2 bound MEC-12/MEC-7 microtubule | | Descriptor: | Alpha-tubulin N-acetyltransferase 2, GUANOSINE-5'-TRIPHOSPHATE, PHOSPHOMETHYLPHOSPHONIC ACID GUANYLATE ESTER, ... | | Authors: | Lam, W.H, Yu, D, Zhai, Y, Ti, S. | | Deposit date: | 2024-02-06 | | Release date: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Tubulin acetyltransferases access and modify the microtubule luminal K40 residue through anchors in taxane-binding pockets.
Nat.Struct.Mol.Biol., 2024
|
|
8YAL
 
 | | ATAT-2 bound K40Q MEC-12/MEC-7 microtubule | | Descriptor: | ACETYL COENZYME *A, Alpha-tubulin N-acetyltransferase 2, GUANOSINE-5'-TRIPHOSPHATE, ... | | Authors: | Lam, W.H, Yu, D, Zhai, Y, Ti, S. | | Deposit date: | 2024-02-09 | | Release date: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Tubulin acetyltransferases access and modify the microtubule luminal K40 residue through anchors in taxane-binding pockets.
Nat.Struct.Mol.Biol., 2024
|
|
7WVM
 
 | | The complex structure of PD-1 and cemiplimab | | Descriptor: | Heavy Chain of Cemiplimab, Light Chain of Cemiplimab, Programmed cell death protein 1 | | Authors: | Lu, D, Xu, Z.P, Liu, K.F, Tan, S.G, Gao, G.F, Chai, Y. | | Deposit date: | 2022-02-10 | | Release date: | 2022-04-20 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (3.4 Å) | | Cite: | PD-1 N58-Glycosylation-Dependent Binding of Monoclonal Antibody Cemiplimab for Immune Checkpoint Therapy.
Front Immunol, 13, 2022
|
|
