4DPG
 
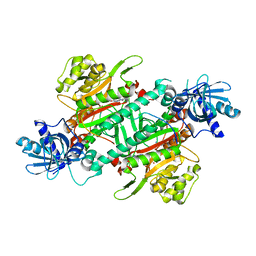 | | Crystal Structure of Human LysRS: P38/AIMP2 Complex I | | Descriptor: | ALANINE, Aminoacyl tRNA synthase complex-interacting multifunctional protein 2, DIPHOSPHOMETHYLPHOSPHONIC ACID ADENOSYL ESTER, ... | | Authors: | Fang, P, Wang, J, Bennett, S.P, Guo, M. | | Deposit date: | 2012-02-13 | | Release date: | 2013-02-13 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.844 Å) | | Cite: | Structural Switch of Lysyl-tRNA Synthetase between Translation and Transcription.
Mol.Cell, 49, 2013
|
|
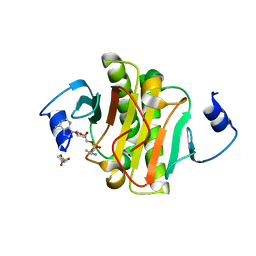
4EQH
 
 | |
4EQE
 
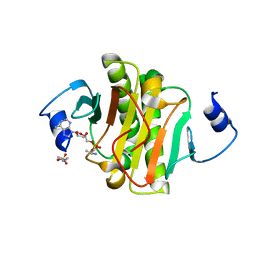 | |
6J5S
 
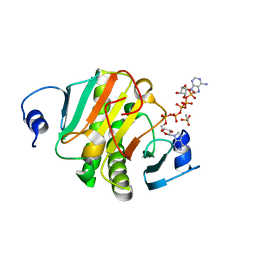 | | Crystal structure of human HINT1 mutant complexing with AP5A | | Descriptor: | BIS(ADENOSINE)-5'-PENTAPHOSPHATE, ETHANESULFONIC ACID, Histidine triad nucleotide-binding protein 1 | | Authors: | Wang, J, Fang, P, Guo, M. | | Deposit date: | 2019-01-11 | | Release date: | 2019-09-25 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.02 Å) | | Cite: | Second messenger Ap4A polymerizes target protein HINT1 to transduce signals in Fc epsilon RI-activated mast cells.
Nat Commun, 10, 2019
|
|
6J53
 
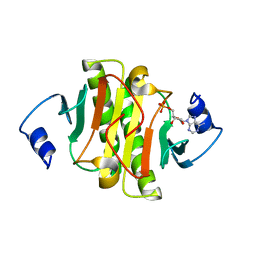 | | Crystal structure of human HINT1 complexing with ATP | | Descriptor: | ADENOSINE MONOPHOSPHATE, Histidine triad nucleotide-binding protein 1 | | Authors: | Wang, J, Fang, P, Guo, M. | | Deposit date: | 2019-01-10 | | Release date: | 2019-09-25 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.52 Å) | | Cite: | Second messenger Ap4A polymerizes target protein HINT1 to transduce signals in Fc epsilon RI-activated mast cells.
Nat Commun, 10, 2019
|
|
6J58
 
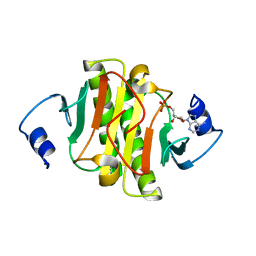 | | Crystal structure of human HINT1 complexing with AP4A | | Descriptor: | ADENOSINE MONOPHOSPHATE, Histidine triad nucleotide-binding protein 1 | | Authors: | Wang, J, Fang, P, Guo, M. | | Deposit date: | 2019-01-10 | | Release date: | 2019-09-25 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.521 Å) | | Cite: | Second messenger Ap4A polymerizes target protein HINT1 to transduce signals in Fc epsilon RI-activated mast cells.
Nat Commun, 10, 2019
|
|
6J64
 
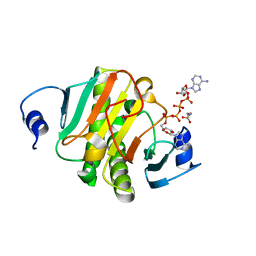 | | Crystal structure of human HINT1 mutant complexing with AP4A | | Descriptor: | 2-AMINOETHANESULFONIC ACID, BIS(ADENOSINE)-5'-TETRAPHOSPHATE, Histidine triad nucleotide-binding protein 1 | | Authors: | Wang, J, Fang, P, Guo, M. | | Deposit date: | 2019-01-14 | | Release date: | 2019-09-25 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (0.95 Å) | | Cite: | Second messenger Ap4A polymerizes target protein HINT1 to transduce signals in Fc epsilon RI-activated mast cells.
Nat Commun, 10, 2019
|
|
6J5Z
 
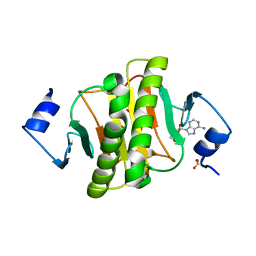 | | Crystal structure of human HINT1 mutant complexing with AP3A | | Descriptor: | ADENOSINE, ETHANESULFONIC ACID, Histidine triad nucleotide-binding protein 1 | | Authors: | Wang, J, Fang, P, Guo, M. | | Deposit date: | 2019-01-12 | | Release date: | 2019-09-25 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Second messenger Ap4A polymerizes target protein HINT1 to transduce signals in Fc epsilon RI-activated mast cells.
Nat Commun, 10, 2019
|
|
6J65
 
 | | Crystal structure of human HINT1 mutant complexing with AP4A II | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, BIS(ADENOSINE)-5'-TETRAPHOSPHATE, Histidine triad nucleotide-binding protein 1 | | Authors: | Wang, J, Fang, P, Guo, M. | | Deposit date: | 2019-01-14 | | Release date: | 2019-09-25 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.42 Å) | | Cite: | Second messenger Ap4A polymerizes target protein HINT1 to transduce signals in Fc epsilon RI-activated mast cells.
Nat Commun, 10, 2019
|
|
3UV9
 
 | | Structure of the rhesus monkey TRIM5alpha deltav1 PRYSPRY domain | | Descriptor: | Tripartite motif-containing protein 5 | | Authors: | Biris, N, Yang, Y, Taylor, A.B, Tomashevskii, A, Guo, M, Hart, P.J, Diaz-Griffero, F, Ivanov, D. | | Deposit date: | 2011-11-29 | | Release date: | 2012-08-08 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.549 Å) | | Cite: | Structure of the rhesus monkey TRIM5alpha PRYSPRY domain, the HIV capsid recognition module.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
8H2I
 
 | | Near-atomic structure of five-fold averaged PBCV-1 capsid | | Descriptor: | MCPv1, MCPv2, MCPv3, ... | | Authors: | Shao, Q, Agarkova, I.V, Noel, E.A, Dunigan, D.D, Liu, Y, Wang, A, Guo, M, Xie, L, Zhao, X, Rossmann, M.G, Van Etten, J.L, Klose, T, Fang, Q. | | Deposit date: | 2022-10-06 | | Release date: | 2022-11-16 | | Last modified: | 2023-09-06 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Near-atomic, non-icosahedrally averaged structure of giant virus Paramecium bursaria chlorella virus 1.
Nat Commun, 13, 2022
|
|
3SJ2
 
 | | A Crystal Structure of a Model of the Repeating r(CGG) Transcript Found in Fragile X Syndrome | | Descriptor: | ACETATE ION, RNA (5'-R(*UP*UP*GP*GP*GP*CP*CP*GP*GP*CP*GP*GP*CP*GP*GP*GP*UP*CP*C)-3'), RNA (5'-R(P*GP*GP*GP*CP*CP*GP*GP*CP*GP*GP*CP*GP*GP*GP*UP*CP*C)-3') | | Authors: | Kumar, A, Pengfei, F, Park, H, Nettles, K, Guo, M, Disney, M.D. | | Deposit date: | 2011-06-20 | | Release date: | 2011-08-03 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.36 Å) | | Cite: | A Crystal Structure of a Model of the Repeating r(CGG) Transcript Found in Fragile X Syndrome.
Chembiochem, 12, 2011
|
|
8HLL
 
 | | Crystal structure of p53/BCL2 fusion complex (complex 1) | | Descriptor: | Apoptosis regulator Bcl-2, Cellular tumor antigen p53, ZINC ION | | Authors: | Wei, H, Guo, M, Wang, H, Chen, Y. | | Deposit date: | 2022-11-30 | | Release date: | 2023-07-26 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2.62 Å) | | Cite: | Structures of p53/BCL-2 complex suggest a mechanism for p53 to antagonize BCL-2 activity.
Nat Commun, 14, 2023
|
|
3SZX
 
 | | Crystal Structure of the Triplet Repeat in Myotonic Dystrophy Reveals Heterogeneous 1x1 Nucleotide UU Internal Loop Conformations | | Descriptor: | RNA (5'-R(P*UP*UP*GP*GP*GP*CP*CP*UP*GP*CP*UP*GP*CP*UP*GP*GP*UP*CP*C)-3') | | Authors: | Kumar, A, Park, H, Pengfei, F, Parkesh, R, Guo, M, Nettles, K.W, Disney, M.D. | | Deposit date: | 2011-07-19 | | Release date: | 2012-04-25 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.204 Å) | | Cite: | Crystal Structure of the Triplet Repeat in Myotonic Dystrophy Reveals Heterogeneous 1x1 Nucleotide UU Internal Loop Conformations
Biochemistry, 50, 2011
|
|
3SYW
 
 | | Crystal Structure of the Triplet Repeat in Myotonic Dystrophy Reveals Heterogeneous 1x1 Nucleotide UU Internal Loop Conformations | | Descriptor: | PHOSPHATE ION, RNA (5'-R(*UP*UP*GP*GP*GP*CP*CP*UP*GP*CP*UP*GP*CP*UP*GP*GP*UP*CP*C)-3') | | Authors: | Kumar, A, Park, H, Pengfei, F, Parkesh, R, Guo, M, Nettles, K.W, Disney, M.D. | | Deposit date: | 2011-07-18 | | Release date: | 2012-04-25 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.57 Å) | | Cite: | Crystal Structure of the Triplet Repeat in Myotonic Dystrophy Reveals Heterogeneous 1x1 Nucleotide UU Internal Loop Conformations
Biochemistry, 50, 2011
|
|
8IQM
 
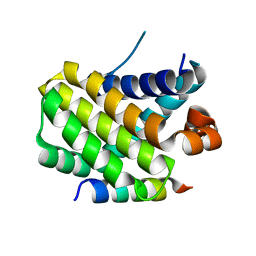 | | Structural basis of the specificity and interaction mechanism of Bmf binding to pro-survival proteins | | Descriptor: | Bcl2 modifying factor, Induced myeloid leukemia cell differentiation protein Mcl-1 | | Authors: | Wang, H, Guo, M, Wei, H, Chen, Y. | | Deposit date: | 2023-03-16 | | Release date: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.967 Å) | | Cite: | Structural basis of the specificity and interaction mechanism of Bmf binding to pro-survival Bcl-2 family proteins.
Comput Struct Biotechnol J, 21, 2023
|
|
8IQK
 
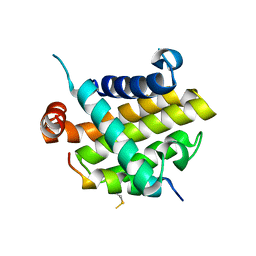 | | Structural basis of the specificity and interaction mechanism of Bmf binding to pro-survival proteins | | Descriptor: | Bcl-2-like protein 1, Bcl-2-modifying factor | | Authors: | Wang, H, Guo, M, Wei, H, Chen, Y. | | Deposit date: | 2023-03-16 | | Release date: | 2023-08-23 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.879 Å) | | Cite: | Structural basis of the specificity and interaction mechanism of Bmf binding to pro-survival Bcl-2 family proteins.
Comput Struct Biotechnol J, 21, 2023
|
|
8IQL
 
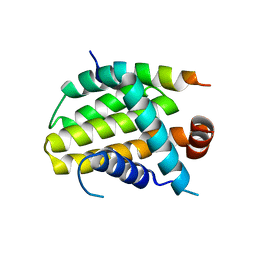 | | Structural basis of the specificity and interaction mechanism of Bmf binding to pro-survival proteins | | Descriptor: | Apoptosis regulator Bcl-2, Bcl-2-modifying factor | | Authors: | Wang, H, Guo, M, Wei, H, Chen, Y. | | Deposit date: | 2023-03-16 | | Release date: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.9577 Å) | | Cite: | Structural basis of the specificity and interaction mechanism of Bmf binding to pro-survival Bcl-2 family proteins.
Comput Struct Biotechnol J, 21, 2023
|
|
5YZX
 
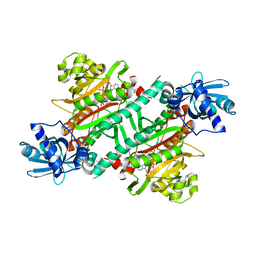 | | Crystal structure of E.coli LysU T146D mutant | | Descriptor: | BIS(ADENOSINE)-5'-TETRAPHOSPHATE, CALCIUM ION, Lysine--tRNA ligase, ... | | Authors: | Fang, P, Guo, M. | | Deposit date: | 2017-12-16 | | Release date: | 2018-12-19 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | A proposed role for MSC to reserve the canonical function in high eukaryotes prior to stimuli
To Be Published
|
|
6AKO
 
 | | Crystal Structure of FOXC2 DBD Bound to DBE2 DNA | | Descriptor: | DNA (5'-D(CP*AP*AP*AP*AP*TP*GP*TP*AP*AP*AP*CP*AP*AP*GP*A)-3'), DNA (5'-D(TP*CP*TP*TP*GP*TP*TP*TP*AP*CP*AP*TP*TP*TP*TP*G)-3'), Forkhead box protein C2, ... | | Authors: | Chen, X, Wei, H, Li, J, Liang, X, Dai, S, Jiang, L, Guo, M, Chen, Y. | | Deposit date: | 2018-09-03 | | Release date: | 2019-02-06 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.396 Å) | | Cite: | Structural basis for DNA recognition by FOXC2.
Nucleic Acids Res., 47, 2019
|
|
6AKP
 
 | | Crystal Structural of FOXC2 DNA binding domain bound to PC promoter | | Descriptor: | DNA (5'-D(AP*CP*AP*CP*AP*AP*AP*TP*AP*TP*TP*TP*GP*TP*GP*T)-3'), Forkhead box protein C2, MAGNESIUM ION | | Authors: | Chen, X, Wei, H, Li, J, Liang, X, Dai, S, Jiang, L, Guo, M, Chen, Y. | | Deposit date: | 2018-09-03 | | Release date: | 2019-02-06 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.323 Å) | | Cite: | Structural basis for DNA recognition by FOXC2.
Nucleic Acids Res., 47, 2019
|
|
5XP5
 
 | | C-Src in complex with ATP-Chf | | Descriptor: | MAGNESIUM ION, Proto-oncogene tyrosine-protein kinase Src, [(2R,3S,4R,5R)-5-(6-aminopurin-9-yl)-3,4-bis(oxidanyl)oxolan-2-yl]methoxy-[(S)-fluoranyl-[oxidanyl(phosphonooxy)phosphoryl]methyl]phosphinic acid | | Authors: | Duan, Y, Guo, M, Dai, S, Chen, L, Chen, Y. | | Deposit date: | 2017-06-01 | | Release date: | 2018-02-07 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.101 Å) | | Cite: | Remarkably Stereospecific Utilization of ATP alpha , beta-Halomethylene Analogues by Protein Kinases.
J. Am. Chem. Soc., 139, 2017
|
|
5XBL
 
 | | Structure of nuclease in complex with associated protein | | Descriptor: | Associated protein, CRISPR-associated endonuclease Cas9/Csn1, RNA (98-MER) | | Authors: | Dong, D, Guo, M, Wang, S, Zhu, Y, Huang, Z. | | Deposit date: | 2017-03-20 | | Release date: | 2017-06-14 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (3.052 Å) | | Cite: | Structural basis of CRISPR-SpyCas9 inhibition by an anti-CRISPR protein
Nature, 546, 2017
|
|
