4KN6
 
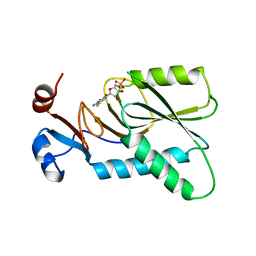 | | Crystal structure of human hypoxanthine-guanine phosphoribosyltransferase in complex with 6-fluoro-3-hydroxy-2-pyrazinecarboxamide (T-705) ribose-5'-monophosphate | | Descriptor: | 6-fluoro-3-oxo-4-(5-O-phosphono-beta-D-ribofuranosyl)-3,4-dihydropyrazine-2-carboxamide, Hypoxanthine-guanine phosphoribosyltransferase | | Authors: | Naesens, L, Guddat, L, Keough, D, van Kuilenburg, A.B.P, Meijer, J, Vande Voorde, J, Balzarini, J. | | Deposit date: | 2013-05-08 | | Release date: | 2013-08-14 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.728 Å) | | Cite: | Role of human hypoxanthine Guanine phosphoribosyltransferase in activation of the antiviral agent T-705 (favipiravir).
Mol.Pharmacol., 84, 2013
|
|
6NC5
 
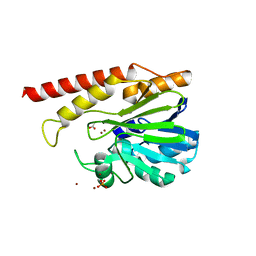 | | Cronobacter sakazakii (Enterobacter sakazakii) Metallo-beta-lactamse HARLDQ motif | | Descriptor: | ACETATE ION, Beta-lactamase, PHOSPHATE ION, ... | | Authors: | Monteiro Pedroso, M, Waite, D, Natasa, M, McGeary, R, Guddat, L, Hugenholtz, P, Schenk, G. | | Deposit date: | 2018-12-10 | | Release date: | 2019-12-25 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.074 Å) | | Cite: | Broad spectrum antibiotic-degrading metallo-beta-lactamases are phylogenetically diverse.
Protein Cell, 11, 2020
|
|
5UQ6
 
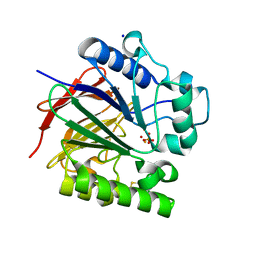 | | PIG PURPLE ACID PHOSPHATASE COMPLEXED WITH PHOSPHATE IN TWO COORDINATION MODES ALONG WITH A BRIDGING HYDROXIDE ION | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, FE (III) ION, HYDROXIDE ION, ... | | Authors: | Selleck, C, Guddat, L, Schenk, G, Clayton, D. | | Deposit date: | 2017-02-06 | | Release date: | 2017-05-24 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.182 Å) | | Cite: | Visualization of the Reaction Trajectory and Transition State in a Hydrolytic Reaction Catalyzed by a Metalloenzyme.
Chemistry, 23, 2017
|
|
8UAJ
 
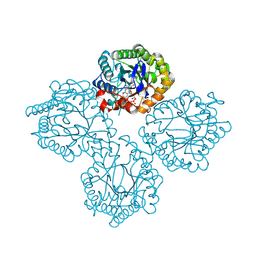 | | Succinate Bound Crystal Structure of Thermus scotoductus SA-01 Ene-reductase | | Descriptor: | FLAVIN MONONUCLEOTIDE, NADPH dehydrogenase, SUCCINIC ACID | | Authors: | Wilson, L.A, Guddat, L, Schenk, G, Scott, C. | | Deposit date: | 2023-09-21 | | Release date: | 2024-07-03 | | Method: | X-RAY DIFFRACTION (2.14 Å) | | Cite: | Structural Characterization of Enzymatic Interactions with Functional Nicotinamide Cofactor Biomimetics
Catalysts, 14, 2024
|
|
5JV5
 
 | |
5KAP
 
 | |
5JSQ
 
 | |
5KAM
 
 | |
5K51
 
 | |
5XKA
 
 | | Crystal structure of M.tuberculosis PknI kinase domain | | Descriptor: | Serine/threonine-protein kinase PknI | | Authors: | Yan, Q, Jiang, D, Qian, L, Zhang, Q, Zhang, W, Zhou, W, Mi, K, Guddat, L, Yang, H, Rao, Z. | | Deposit date: | 2017-05-06 | | Release date: | 2018-05-16 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.599 Å) | | Cite: | Structural Insight into the Activation of PknI Kinase from M. tuberculosis via Dimerization of the Extracellular Sensor Domain.
Structure, 25, 2017
|
|
6MFI
 
 | | MIM-2 Metallo-Beta-Lactamase | | Descriptor: | Metallo-beta-lactamase, ZINC ION | | Authors: | Selleck, C, Guddat, L, Schenk, G, Monteiro Pedroso, M. | | Deposit date: | 2018-09-11 | | Release date: | 2019-11-13 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.839 Å) | | Cite: | characterization of the B3 MBLs MIM-1 and MIM-2 from environmental microorganisms.
Chemistry, 23, 2017
|
|
6MXC
 
 | |
6MXB
 
 | |
6MXG
 
 | |
6MXD
 
 | |
6DR8
 
 | | Metallo-beta-lactamase from Cronobacter sakazakii (Enterobacter sakazakii) HARLDQ motif mutant S60/R118H/Q121H/K254H | | Descriptor: | (2-hydroxyethoxy)acetaldehyde, Beta-lactamase, PHOSPHATE ION, ... | | Authors: | Monteiro Pedroso, M, Waite, D, Natasa, M, McGeary, R, Guddat, L, Hugenholtz, P, Schenk, G. | | Deposit date: | 2018-06-11 | | Release date: | 2019-06-19 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.476 Å) | | Cite: | Broad spectrum antibiotic-degrading metallo-beta-lactamases are phylogenetically diverse
Protein Cell, 2020
|
|
6DQH
 
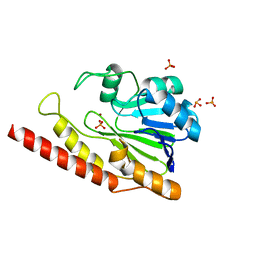 | | Cronobacter sakazakii (Enterobacter sakazakii) Metallo-beta-lactamse HARLDQ motif | | Descriptor: | Beta-lactamase, PHOSPHATE ION, ZINC ION | | Authors: | Monteiro Pedroso, M, Waite, D, Natasa, M, McGeary, R, Guddat, L, Hugenholtz, P, Schenk, G. | | Deposit date: | 2018-06-11 | | Release date: | 2019-06-19 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.104 Å) | | Cite: | Broad spectrum antibiotic-degrading metallo-beta-lactamases are phylogenetically diverse
Protein Cell, 2020
|
|
6DQ2
 
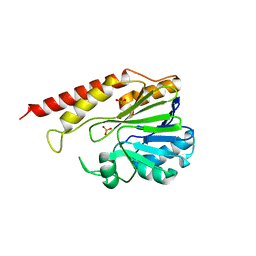 | | Cronobacter sakazakii (Enterobacter sakazakii) Metallo-beta-lactamse HARLDQ motif mutant S60 | | Descriptor: | Beta-lactamase, PHOSPHATE ION | | Authors: | Monteiro Pedroso, M, Waite, D, Natasa, M, McGeary, R, Guddat, L, Hugenholtz, P, Schenk, G. | | Deposit date: | 2018-06-10 | | Release date: | 2019-06-12 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.158 Å) | | Cite: | Broad spectrum antibiotic-degrading metallo-beta-lactamases are phylogenetically diverse
Protein Cell, 2020
|
|
6DN4
 
 | | Cronobacter sakazakii (Enterobacter sakazakii) Metallo-beta-lactamse HARLDQ motif | | Descriptor: | Beta-lactamase | | Authors: | Monteiro Pedroso, M, Waite, D, Natasa, M, McGeary, R, Guddat, L, Hugenholtz, P, Schenk, G. | | Deposit date: | 2018-06-06 | | Release date: | 2019-06-12 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.99 Å) | | Cite: | Broad spectrum antibiotic-degrading metallo-beta-lactamases are phylogenetically diverse
Protein Cell, 2020
|
|
6APU
 
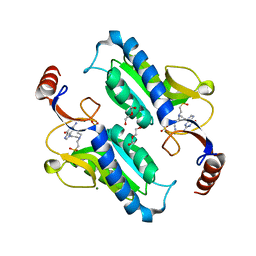 | | Crystal structure of Trypanosoma brucei hypoxanthine-guanine phosphoribosyltranferase in complex with (2-{[2-(2-amino-6-oxo-1,6-dihydro-9H-purin-9-yl)ethyl](3-aminopropyl)amino}ethyl)phosphonic acid | | Descriptor: | (2-{[2-(2-amino-6-oxo-1,6-dihydro-9H-purin-9-yl)ethyl](3-aminopropyl)amino}ethyl)phosphonic acid, DI(HYDROXYETHYL)ETHER, Hypoxanthine-guanine phosphoribosyltransferase, ... | | Authors: | Teran, D, Guddat, L. | | Deposit date: | 2017-08-18 | | Release date: | 2018-03-14 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.842 Å) | | Cite: | Evaluation of the Trypanosoma brucei 6-oxopurine salvage pathway as a potential target for drug discovery.
PLoS Negl Trop Dis, 12, 2018
|
|
6KGU
 
 | | Crystal structure of Penicillin binding protein 3 (PBP3) from Mycobacterium tuerculosis, complexed with aztreonam | | Descriptor: | 2-({[(1Z)-1-(2-amino-1,3-thiazol-4-yl)-2-oxo-2-{[(2S,3S)-1-oxo-3-(sulfoamino)butan-2-yl]amino}ethylidene]amino}oxy)-2-methylpropanoic acid, COBALT (II) ION, Penicillin-binding protein PbpB | | Authors: | Lu, Z.K, Zhang, A.L, Liu, X, Guddat, L, Yang, H.T, Rao, Z.H. | | Deposit date: | 2019-07-12 | | Release date: | 2020-03-11 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.106 Å) | | Cite: | Structures ofMycobacterium tuberculosisPenicillin-Binding Protein 3 in Complex with Fivebeta-Lactam Antibiotics Reveal Mechanism of Inactivation.
Mol.Pharmacol., 97, 2020
|
|
6KGV
 
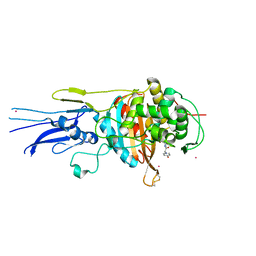 | | Crystal structure of Penicillin binding protein 3 (PBP3) from Mycobacterium tuerculosis, complexed with amoxicillin | | Descriptor: | 2-{1-[2-AMINO-2-(4-HYDROXY-PHENYL)-ACETYLAMINO]-2-OXO-ETHYL}-5,5-DIMETHYL-THIAZOLIDINE-4-CARBOXYLIC ACID, COBALT (II) ION, Penicillin-binding protein PbpB | | Authors: | Lu, Z.K, Zhang, A.L, Liu, X, Guddat, L, Yang, H.T, Rao, Z.H. | | Deposit date: | 2019-07-12 | | Release date: | 2020-03-11 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.301 Å) | | Cite: | Structures ofMycobacterium tuberculosisPenicillin-Binding Protein 3 in Complex with Fivebeta-Lactam Antibiotics Reveal Mechanism of Inactivation.
Mol.Pharmacol., 97, 2020
|
|
6KGH
 
 | | Crystal structure of Penicillin binding protein 3 (PBP3) from Mycobacterium tuerculosis (apo-form) | | Descriptor: | COBALT (II) ION, Penicillin-binding protein PbpB, SODIUM ION | | Authors: | Lu, Z.K, Zhang, A.L, Liu, X, Guddat, L, Yang, H.T, Rao, Z.H. | | Deposit date: | 2019-07-11 | | Release date: | 2020-03-11 | | Method: | X-RAY DIFFRACTION (2.108 Å) | | Cite: | Structures ofMycobacterium tuberculosisPenicillin-Binding Protein 3 in Complex with Fivebeta-Lactam Antibiotics Reveal Mechanism of Inactivation.
Mol.Pharmacol., 97, 2020
|
|
6KGS
 
 | | Crystal structure of Penicillin binding protein 3 (PBP3) from Mycobacterium tuerculosis, complexed with meropenem | | Descriptor: | (4R,5S)-3-{[(3S,5S)-5-(dimethylcarbamoyl)pyrrolidin-3-yl]sulfanyl}-5-[(2S,3R)-3-hydroxy-1-oxobutan-2-yl]-4-methyl-4,5-d ihydro-1H-pyrrole-2-carboxylic acid, COBALT (II) ION, Penicillin-binding protein PbpB | | Authors: | Lu, Z.K, Zhang, A.L, Liu, X, Guddat, L, Yang, H.T, Rao, Z.H. | | Deposit date: | 2019-07-12 | | Release date: | 2020-03-11 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.309 Å) | | Cite: | Structures ofMycobacterium tuberculosisPenicillin-Binding Protein 3 in Complex with Fivebeta-Lactam Antibiotics Reveal Mechanism of Inactivation.
Mol.Pharmacol., 97, 2020
|
|
6KGW
 
 | | Crystal structure of Penicillin binding protein 3 (PBP3) from Mycobacterium tuerculosis, complexed with ampicillin | | Descriptor: | (2R,4S)-2-[(1R)-1-{[(2R)-2-amino-2-phenylacetyl]amino}-2-oxoethyl]-5,5-dimethyl-1,3-thiazolidine-4-carboxylic acid, COBALT (II) ION, Penicillin-binding protein PbpB | | Authors: | Lu, Z.K, Zhang, A.L, Liu, X, Guddat, L, Yang, H.T, Rao, Z.H. | | Deposit date: | 2019-07-12 | | Release date: | 2020-03-11 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.407 Å) | | Cite: | Structures ofMycobacterium tuberculosisPenicillin-Binding Protein 3 in Complex with Fivebeta-Lactam Antibiotics Reveal Mechanism of Inactivation.
Mol.Pharmacol., 97, 2020
|
|
