1TFG
 
 | |
2DGK
 
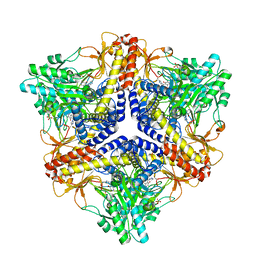 | | Crystal structure of an N-terminal deletion mutant of Escherichia coli GadB in an autoinhibited state (aldamine) | | Descriptor: | 1,2-ETHANEDIOL, Glutamate decarboxylase beta, PYRIDOXAL-5'-PHOSPHATE, ... | | Authors: | Gruetter, M.G, Capitani, G, Gut, H. | | Deposit date: | 2006-03-14 | | Release date: | 2006-06-20 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Escherichia coli acid resistance: pH-sensing, activation by chloride and autoinhibition in GadB
Embo J., 25, 2006
|
|
2DGL
 
 | | Crystal structure of Escherichia coli GadB in complex with bromide | | Descriptor: | ACETIC ACID, BROMIDE ION, Glutamate decarboxylase beta, ... | | Authors: | Gruetter, M.G, Capitani, G, Gut, H. | | Deposit date: | 2006-03-14 | | Release date: | 2006-06-20 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (3.15 Å) | | Cite: | Escherichia coli acid resistance: pH-sensing, activation by chloride and autoinhibition in GadB
Embo J., 25, 2006
|
|
2DGM
 
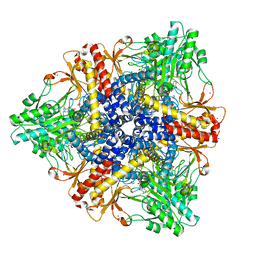 | | Crystal structure of Escherichia coli GadB in complex with iodide | | Descriptor: | ACETIC ACID, DI(HYDROXYETHYL)ETHER, FORMIC ACID, ... | | Authors: | Gruetter, M.G, Capitani, G, Gut, H. | | Deposit date: | 2006-03-14 | | Release date: | 2006-06-20 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Escherichia coli acid resistance: pH-sensing, activation by chloride and autoinhibition in GadB
Embo J., 25, 2006
|
|
1RNE
 
 | |
1SBN
 
 | |
1SIB
 
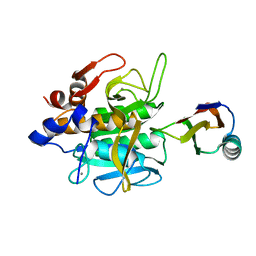 | |
2WL1
 
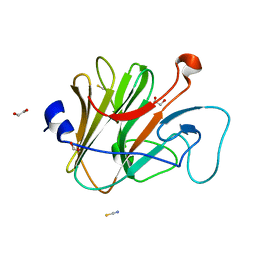 | | Pyrin PrySpry domain | | Descriptor: | 1,2-ETHANEDIOL, PYRIN, THIOCYANATE ION | | Authors: | Weinert, C, Mittl, P.R, Gruetter, M.G. | | Deposit date: | 2009-06-19 | | Release date: | 2009-10-20 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | The Crystal Structure of Human Pyrin B30.2 Domain: Implications for Mutations Associated with Familial Mediterranean Fever.
J.Mol.Biol., 394, 2009
|
|
1AIE
 
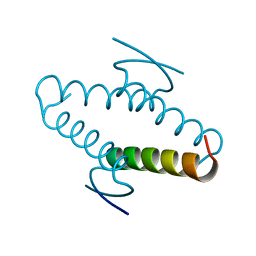 | |
2V50
 
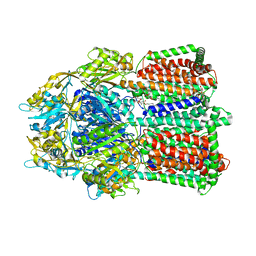 | |
1BX7
 
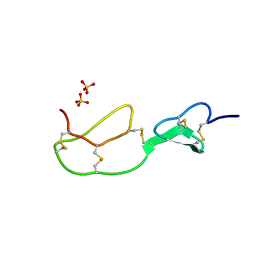 | | HIRUSTASIN FROM HIRUDO MEDICINALIS AT 1.2 ANGSTROMS | | Descriptor: | HIRUSTASIN, SULFATE ION | | Authors: | Uson, I, Sheldrick, G.M, De La Fortelle, E, Bricogne, G, Di Marco, S, Priestle, J.P, Gruetter, M.G, Mittl, P.R.E. | | Deposit date: | 1998-10-14 | | Release date: | 1999-04-27 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | The 1.2 A crystal structure of hirustasin reveals the intrinsic flexibility of a family of highly disulphide-bridged inhibitors.
Structure Fold.Des., 7, 1999
|
|
1BX8
 
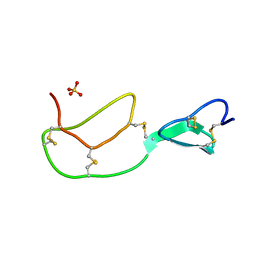 | | HIRUSTASIN FROM HIRUDO MEDICINALIS AT 1.4 ANGSTROMS | | Descriptor: | HIRUSTASIN, SULFATE ION | | Authors: | Uson, I, Sheldrick, G.M, De La Fortelle, E, Bricogne, G, Di Marco, S, Priestle, J.P, Gruetter, M.G, Mittl, P.R.E. | | Deposit date: | 1998-10-14 | | Release date: | 1999-04-27 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | The 1.2 A crystal structure of hirustasin reveals the intrinsic flexibility of a family of highly disulphide-bridged inhibitors.
Structure Fold.Des., 7, 1999
|
|
2FBE
 
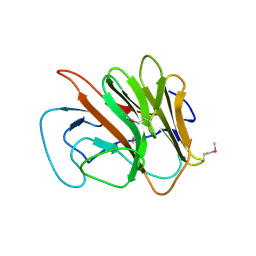 | | Crystal Structure of the PRYSPRY-domain | | Descriptor: | PREDICTED: similar to ret finger protein-like 1 | | Authors: | Gruetter, C, Briand, C, Capitani, G, Mittl, P.R, Gruetter, M.G. | | Deposit date: | 2005-12-09 | | Release date: | 2006-01-10 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.52 Å) | | Cite: | Structure of the PRYSPRY-domain: Implications for autoinflammatory diseases
Febs Lett., 580, 2006
|
|
1C94
 
 | | REVERSING THE SEQUENCE OF THE GCN4 LEUCINE ZIPPER DOES NOT AFFECT ITS FOLD. | | Descriptor: | RETRO-GCN4 LEUCINE ZIPPER | | Authors: | Mittl, P.R.E, Deillon, C.A, Sargent, D, Liu, N, Klauser, S, Thomas, R.M, Gutte, B, Gruetter, M.G. | | Deposit date: | 1999-07-30 | | Release date: | 2000-03-22 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.08 Å) | | Cite: | The retro-GCN4 leucine zipper sequence forms a stable three-dimensional structure.
Proc.Natl.Acad.Sci.USA, 97, 2000
|
|
3EO0
 
 | |
5FIO
 
 | | DARPins as a new tool for experimental phasing in protein crystallography | | Descriptor: | MALTOSE-BINDING PERIPLASMIC PROTEIN, MERCURY (II) ION, NI3C DARPIN MUTANT5 HG-SITE N1 | | Authors: | Batyuk, A, Honegger, A, Andres, F, Briand, C, Gruetter, M, Plueckthun, A. | | Deposit date: | 2015-09-30 | | Release date: | 2016-11-16 | | Last modified: | 2019-10-23 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Darpins as a New Tool for Experimental Phasing in Protein Crystallography
To be Published
|
|
5FIN
 
 | | DARPins as a new tool for experimental phasing in protein crystallography | | Descriptor: | MERCURY (II) ION, NI3C DARPIN MUTANT5 HG-SITE N1 | | Authors: | Batyuk, A, Honegger, A, Andres, F, Briand, C, Gruetter, M, Plueckthun, A. | | Deposit date: | 2015-09-30 | | Release date: | 2016-11-16 | | Last modified: | 2017-07-05 | | Method: | X-RAY DIFFRACTION (2.34 Å) | | Cite: | Darpins as a New Tool for Experimental Phasing in Protein Crystallography
To be Published
|
|
2I1B
 
 | |
3EO1
 
 | |
4HJ0
 
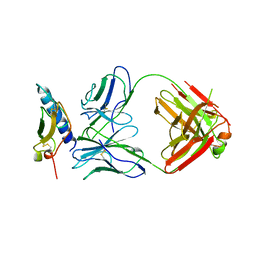 | | Crystal structure of the human GIPr ECD in complex with Gipg013 Fab at 3-A resolution | | Descriptor: | Gastric inhibitory polypeptide receptor, Gipg013 Fab, Antagonizing antibody to the GIP Receptor, ... | | Authors: | Madhurantakam, C, Ravn, P, Gruetter, M.G, Jackson, R.H. | | Deposit date: | 2012-10-12 | | Release date: | 2013-05-29 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structural and Pharmacological Characterization of Novel Potent and Selective Monoclonal Antibody Antagonists of Glucose-dependent Insulinotropic Polypeptide Receptor.
J.Biol.Chem., 288, 2013
|
|
1SVX
 
 | | Crystal structure of a designed selected Ankyrin Repeat protein in complex with the Maltose Binding Protein | | Descriptor: | Ankyrin Repeat Protein off7, Maltose-binding periplasmic protein | | Authors: | Binz, H.K, Amstutz, P, Kohl, A, Stumpp, M.T, Briand, C, Forrer, P, Gruetter, M.G, Plueckthun, A. | | Deposit date: | 2004-03-30 | | Release date: | 2004-05-25 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.24 Å) | | Cite: | High-affinity binders selected from designed ankyrin repeat protein libraries
NAT.BIOTECHNOL., 22, 2004
|
|
154L
 
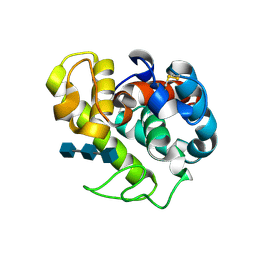 | |
153L
 
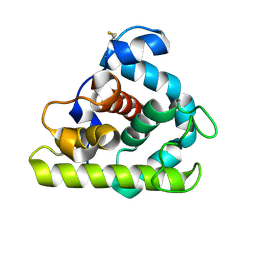 | |
2RKO
 
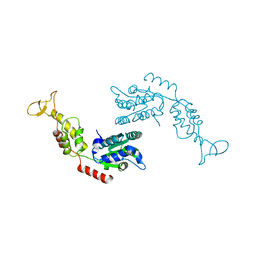 | | Crystal Structure of the Vps4p-dimer | | Descriptor: | Vacuolar protein sorting-associated protein 4 | | Authors: | Hartmann, C, Gruetter, M.G. | | Deposit date: | 2007-10-17 | | Release date: | 2008-02-26 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (3.35 Å) | | Cite: | Vacuolar protein sorting: two different functional states of the AAA-ATPase Vps4p
J.Mol.Biol., 377, 2008
|
|
2V7N
 
 | | Unusual twinning in crystals of the CitS binding antibody Fab fragment f3p4 | | Descriptor: | IMMUNOGLOBULIN HEAVY CHAIN, IMMUNOGLOBULIN LIGHT CHAIN | | Authors: | Frey, D, Huber, T, Plueckthun, A, Gruetter, M.G. | | Deposit date: | 2007-07-31 | | Release date: | 2008-06-17 | | Last modified: | 2019-05-22 | | Method: | X-RAY DIFFRACTION (1.92 Å) | | Cite: | Structure of the Recombinant Antibody Fab Fragment F3P4.
Acta Crystallogr.,Sect.D, 64, 2008
|
|
