1S80
 
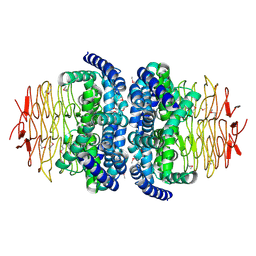 | | Structure of Serine Acetyltransferase from Haemophilis influenzae Rd | | Descriptor: | Serine acetyltransferase | | Authors: | Gorman, J, Gogos, A, Shapiro, L, Burley, S.K, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2004-01-30 | | Release date: | 2004-08-31 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structure of serine acetyltransferase from Haemophilus influenzae Rd.
Acta Crystallogr.,Sect.D, 60, 2004
|
|
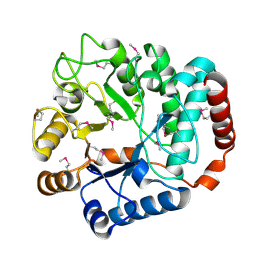
1TR9
 
 | |
1R3D
 
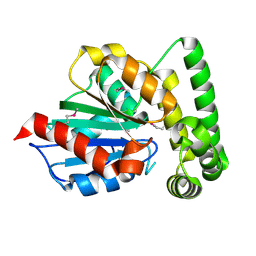 | |
1TVL
 
 | |
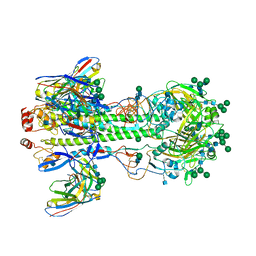
7KC1
 
 | |
7KNI
 
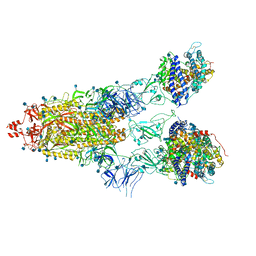 | | Cryo-EM structure of Triple ACE2-bound SARS-CoV-2 Trimer Spike at pH 5.5 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Angiotensin-converting enzyme 2, ... | | Authors: | Gorman, J, Rapp, M, Kwong, P.D, Shapiro, L. | | Deposit date: | 2020-11-04 | | Release date: | 2020-12-16 | | Last modified: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (3.91 Å) | | Cite: | Cryo-EM Structures of SARS-CoV-2 Spike without and with ACE2 Reveal a pH-Dependent Switch to Mediate Endosomal Positioning of Receptor-Binding Domains.
Cell Host Microbe, 28, 2020
|
|
7KNE
 
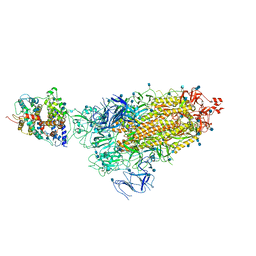 | | Cryo-EM structure of single ACE2-bound SARS-CoV-2 trimer spike at pH 5.5 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Angiotensin-converting enzyme 2, ... | | Authors: | Gorman, J, Rapp, M, Kwong, P.D, Shapiro, L. | | Deposit date: | 2020-11-04 | | Release date: | 2020-12-16 | | Last modified: | 2021-12-15 | | Method: | ELECTRON MICROSCOPY (3.85 Å) | | Cite: | Cryo-EM Structures of SARS-CoV-2 Spike without and with ACE2 Reveal a pH-Dependent Switch to Mediate Endosomal Positioning of Receptor-Binding Domains.
Cell Host Microbe, 28, 2020
|
|
7KMB
 
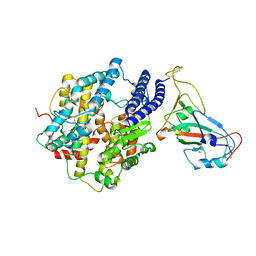 | | ACE2-RBD Focused Refinement Using Symmetry Expansion of Applied C3 for Triple ACE2-bound SARS-CoV-2 Trimer Spike at pH 7.4 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Angiotensin-converting enzyme 2, ... | | Authors: | Gorman, J, Kwong, P.D, Shapiro, L. | | Deposit date: | 2020-11-02 | | Release date: | 2020-12-09 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (3.39 Å) | | Cite: | Cryo-EM Structures of SARS-CoV-2 Spike without and with ACE2 Reveal a pH-Dependent Switch to Mediate Endosomal Positioning of Receptor-Binding Domains.
Cell Host Microbe, 28, 2020
|
|
7KNH
 
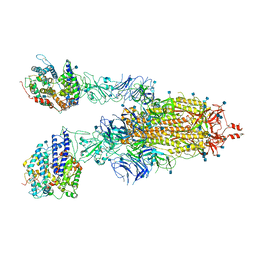 | | Cryo-EM Structure of Double ACE2-Bound SARS-CoV-2 Trimer Spike at pH 5.5 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Angiotensin-converting enzyme 2, ... | | Authors: | Gorman, J, Rapp, M, Kwong, P.D, Shapiro, L. | | Deposit date: | 2020-11-04 | | Release date: | 2020-12-16 | | Last modified: | 2021-12-15 | | Method: | ELECTRON MICROSCOPY (3.74 Å) | | Cite: | Cryo-EM Structures of SARS-CoV-2 Spike without and with ACE2 Reveal a pH-Dependent Switch to Mediate Endosomal Positioning of Receptor-Binding Domains.
Cell Host Microbe, 28, 2020
|
|
7KNB
 
 | | Cryo-EM structure of single ACE2-bound SARS-CoV-2 trimer spike at pH 7.4 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Angiotensin-converting enzyme 2, ... | | Authors: | Gorman, J, Kwong, P.D, Shapiro, L. | | Deposit date: | 2020-11-04 | | Release date: | 2020-12-09 | | Last modified: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (3.93 Å) | | Cite: | Cryo-EM Structures of SARS-CoV-2 Spike without and with ACE2 Reveal a pH-Dependent Switch to Mediate Endosomal Positioning of Receptor-Binding Domains.
Cell Host Microbe, 28, 2020
|
|
7KMS
 
 | | Cryo-EM structure of triple ACE2-bound SARS-CoV-2 trimer spike at pH 7.4 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Angiotensin-converting enzyme 2, ... | | Authors: | Gorman, J, Kwong, P.D, Shapiro, L. | | Deposit date: | 2020-11-03 | | Release date: | 2020-12-09 | | Last modified: | 2021-12-15 | | Method: | ELECTRON MICROSCOPY (3.64 Å) | | Cite: | Cryo-EM Structures of SARS-CoV-2 Spike without and with ACE2 Reveal a pH-Dependent Switch to Mediate Endosomal Positioning of Receptor-Binding Domains.
Cell Host Microbe, 28, 2020
|
|
7KMZ
 
 | | Cryo-EM structure of double ACE2-bound SARS-CoV-2 trimer Spike at pH 7.4 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Angiotensin-converting enzyme 2, ... | | Authors: | Gorman, J, Kwong, P.D, Shapiro, L. | | Deposit date: | 2020-11-03 | | Release date: | 2020-12-09 | | Last modified: | 2021-12-15 | | Method: | ELECTRON MICROSCOPY (3.62 Å) | | Cite: | Cryo-EM Structures of SARS-CoV-2 Spike without and with ACE2 Reveal a pH-Dependent Switch to Mediate Endosomal Positioning of Receptor-Binding Domains.
Cell Host Microbe, 28, 2020
|
|
7L0L
 
 | | Cryo-EM structure of the VRC316 clinical trial, vaccine-elicited, human antibody 316-310-1B11 in complex with an H2 CAN05 HA trimer | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 316-310-1B11 Heavy Chain, 316-310-1B11 Light Chain, ... | | Authors: | Gorman, J, Kwong, P.D. | | Deposit date: | 2020-12-11 | | Release date: | 2021-11-03 | | Last modified: | 2022-03-09 | | Method: | ELECTRON MICROSCOPY (2.85 Å) | | Cite: | A single residue in influenza virus H2 hemagglutinin enhances the breadth of the B cell response elicited by H2 vaccination.
Nat Med, 28, 2022
|
|
7LQV
 
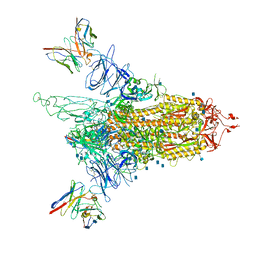 | | Cryo-EM structure of NTD-directed neutralizing antibody 4-8 Fab in complex with SARS-CoV-2 S2P spike | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 4-8 Heavy Chain, 4-8 Light chain, ... | | Authors: | Gorman, J, Rapp, M, Kwong, P.D, Shapiro, L. | | Deposit date: | 2021-02-15 | | Release date: | 2021-03-24 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (3.25 Å) | | Cite: | Potent SARS-CoV-2 Neutralizing Antibodies Directed Against Spike N-Terminal Domain Target a Single Supersite
Cell Host Microbe, 2021
|
|
7LQW
 
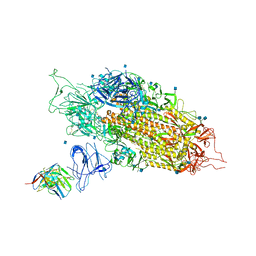 | | Cryo-EM structure of NTD-directed neutralizing antibody 2-17 Fab in complex with SARS-CoV-2 S2P spike | | Descriptor: | 2-17 Heavy Chain, 2-17 Light Chain, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Gorman, J, Rapp, M, Kwong, P.D, Shapiro, L. | | Deposit date: | 2021-02-15 | | Release date: | 2021-03-24 | | Last modified: | 2021-05-26 | | Method: | ELECTRON MICROSCOPY (4.47 Å) | | Cite: | Potent SARS-CoV-2 neutralizing antibodies directed against spike N-terminal domain target a single supersite.
Cell Host Microbe, 29, 2021
|
|
7LZE
 
 | | Cryo-EM Structure of disulfide stabilized HMPV F v4-B | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Fusion glycoprotein F0 | | Authors: | Gorman, J, Kwong, P.D. | | Deposit date: | 2021-03-09 | | Release date: | 2021-08-25 | | Last modified: | 2021-10-06 | | Method: | ELECTRON MICROSCOPY (3.23 Å) | | Cite: | Interprotomer disulfide-stabilized variants of the human metapneumovirus fusion glycoprotein induce high titer-neutralizing responses.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
7LY9
 
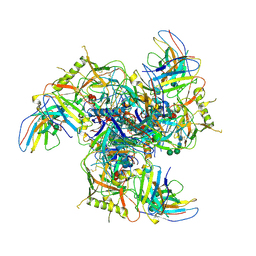 | |
7KJP
 
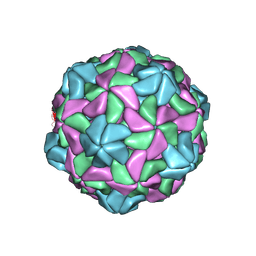 | | Disulfide Stabilized Norovirus GI.1 VLP Shell Region | | Descriptor: | Capsid protein VP1 | | Authors: | Gorman, J, Kwong, P.D. | | Deposit date: | 2020-10-26 | | Release date: | 2020-12-02 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (3.86 Å) | | Cite: | Disulfide stabilization of human norovirus GI.1 virus-like particles focuses immune response toward blockade epitopes.
NPJ Vaccines, 5, 2020
|
|
1Q7E
 
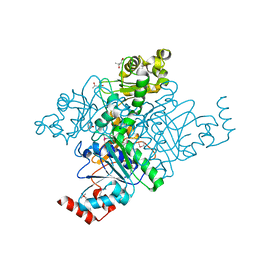 | | Crystal Structure of YfdW protein from E. coli | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, Hypothetical protein yfdW, METHIONINE | | Authors: | Gogos, A, Gorman, J, Shapiro, L, Burley, S.K, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2003-08-18 | | Release date: | 2003-09-30 | | Last modified: | 2021-02-03 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structure of Escherichia coli YfdW, a type III CoA transferase.
Acta Crystallogr.,Sect.D, 60, 2004
|
|
5MP6
 
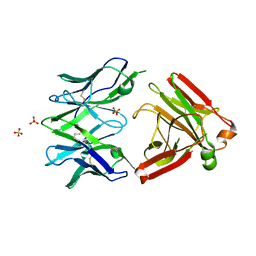 | | Structure of the Unliganded Fab from HIV-1 Neutralizing Antibody CAP248-2B that Binds to the gp120 C-terminus - gp41 Interface, at two Angstrom resolution. | | Descriptor: | CAP248-2B Heavy Chain, CAP248-2B Light Chain, SULFATE ION | | Authors: | Wibmer, C.K, Gorman, J, Kwong, P.D. | | Deposit date: | 2016-12-15 | | Release date: | 2016-12-28 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.959 Å) | | Cite: | Structure and Recognition of a Novel HIV-1 gp120-gp41 Interface Antibody that Caused MPER Exposure through Viral Escape.
PLoS Pathog., 13, 2017
|
|
5T33
 
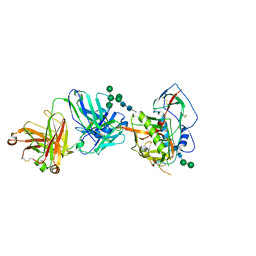 | | Crystal structure of strain-specific glycan-dependent CD4 binding site-directed neutralizing antibody CAP257-RH1, in complex with HIV-1 strain RHPA gp120 core with an oligomannose N276 glycan. | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CAP257-RH1 heavy chain, ... | | Authors: | Wibmer, C.K, Gorman, J, Kwong, P.D. | | Deposit date: | 2016-08-24 | | Release date: | 2016-09-07 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (3.2092 Å) | | Cite: | Structure of an N276-Dependent HIV-1 Neutralizing Antibody Targeting a Rare V5 Glycan Hole Adjacent to the CD4 Binding Site.
J.Virol., 90, 2016
|
|
5L03
 
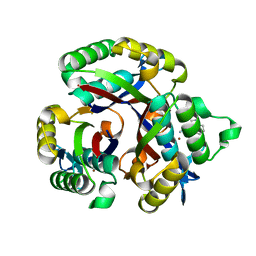 | | Crystal structure of 2-C-METHYL-D-ERYTHRITOL 2,4-CYCLODIPHOSPHATE Synthase from BURKHOLDERIA PSEUDOMALLEI bound to L-tryptophan hydroxamate | | Descriptor: | 2-C-methyl-D-erythritol 2,4-cyclodiphosphate synthase, N-hydroxy-L-tryptophanamide, ZINC ION | | Authors: | Blain, J.M, Ghose, D, Gorman, J.L, Goshu, G.M, Ranieri, G, Zhao, L, Bode, B, Meganathan, R, Walter, R.L, Hagen, T.J, Horn, J.R. | | Deposit date: | 2016-07-26 | | Release date: | 2017-11-08 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.469 Å) | | Cite: | Synthesis and Characterization of the Burkholderia pseudomallei IspF Inhibitor L-tryptophan hydroxamate
To Be Published
|
|
8DW2
 
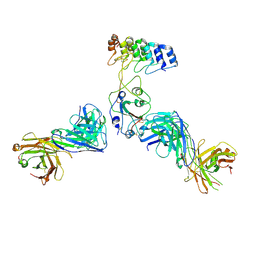 | | Cryo-EM structure of SARS-CoV-2 RBD in complex with anti-SARS-CoV-2 DARPin,SR22, and two antibody Fabs, S309 and CR3022 | | Descriptor: | Antibody CR3022 heavy chain, Antibody CR3022 light chain, Antibody S309 heavy chain, ... | | Authors: | Kwon, Y.D, Gorman, J, Kwong, P.D. | | Deposit date: | 2022-07-30 | | Release date: | 2022-12-07 | | Last modified: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (4.11 Å) | | Cite: | A potent and broad neutralization of SARS-CoV-2 variants of concern by DARPins.
Nat.Chem.Biol., 19, 2023
|
|
8DW3
 
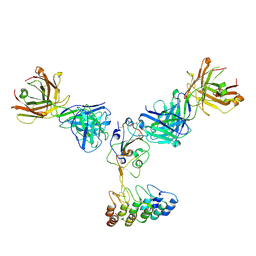 | | Cryo-EM structure of SARS-CoV-2 RBD in complex with anti-SARS-CoV-2 DARPin,SR16m, and two antibody Fabs, S309 and CR3022 | | Descriptor: | Anti-SARS-CoV-2 DARPin SR16m, Antibody S309 light chain, Spike protein S1, ... | | Authors: | Kwon, Y.D, Gorman, J, Kwong, P.D. | | Deposit date: | 2022-07-30 | | Release date: | 2022-12-07 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (4.26 Å) | | Cite: | A potent and broad neutralization of SARS-CoV-2 variants of concern by DARPins.
Nat.Chem.Biol., 19, 2023
|
|
8VUE
 
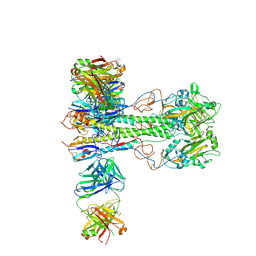 | | L5A7 Fab bound to Indonesia2005 Hemagglutinin | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Hemagglutinin HA1 chain, Hemagglutinin HA2 chain, ... | | Authors: | Olia, A.S, Gorman, J, Kwong, P.D. | | Deposit date: | 2024-01-29 | | Release date: | 2024-05-22 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (3.59 Å) | | Cite: | Anti-idiotype isolation of a broad and potent influenza A virus-neutralizing human antibody.
Front Immunol, 15, 2024
|
|
