5L03
 
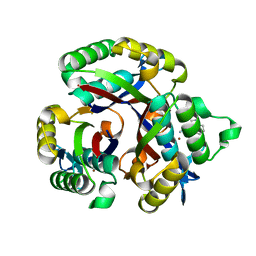 | | Crystal structure of 2-C-METHYL-D-ERYTHRITOL 2,4-CYCLODIPHOSPHATE Synthase from BURKHOLDERIA PSEUDOMALLEI bound to L-tryptophan hydroxamate | | 分子名称: | 2-C-methyl-D-erythritol 2,4-cyclodiphosphate synthase, N-hydroxy-L-tryptophanamide, ZINC ION | | 著者 | Blain, J.M, Ghose, D, Gorman, J.L, Goshu, G.M, Ranieri, G, Zhao, L, Bode, B, Meganathan, R, Walter, R.L, Hagen, T.J, Horn, J.R. | | 登録日 | 2016-07-26 | | 公開日 | 2017-11-08 | | 最終更新日 | 2024-03-06 | | 実験手法 | X-RAY DIFFRACTION (1.469 Å) | | 主引用文献 | Synthesis and Characterization of the Burkholderia pseudomallei IspF Inhibitor L-tryptophan hydroxamate
To Be Published
|
|
8G9X
 
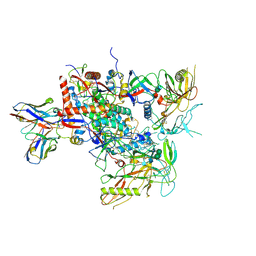 | | Cryo-EM structure of vFP49.02 Fab in complex with HIV-1 Env BG505 DS-SOSIP.664 (conformation 2) | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Envelope glycoprotein gp120, ... | | 著者 | Changela, A, Gorman, J, Kwong, P.D. | | 登録日 | 2023-02-22 | | 公開日 | 2023-04-19 | | 最終更新日 | 2023-06-14 | | 実験手法 | ELECTRON MICROSCOPY (4.46 Å) | | 主引用文献 | Diverse Murine Vaccinations Reveal Distinct Antibody Classes to Target Fusion Peptide and Variation in Peptide Length to Improve HIV Neutralization.
J.Virol., 97, 2023
|
|
8G9W
 
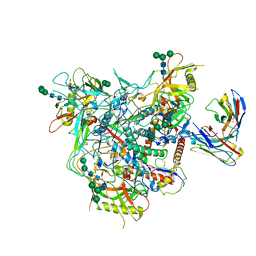 | | Cryo-EM structure of vFP49.02 Fab in complex with HIV-1 Env BG505 DS-SOSIP.664 (conformation 1) | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Envelope glycoprotein gp120, ... | | 著者 | Changela, A, Gorman, J, Kwong, P.D. | | 登録日 | 2023-02-22 | | 公開日 | 2023-04-19 | | 最終更新日 | 2023-06-14 | | 実験手法 | ELECTRON MICROSCOPY (4.66 Å) | | 主引用文献 | Diverse Murine Vaccinations Reveal Distinct Antibody Classes to Target Fusion Peptide and Variation in Peptide Length to Improve HIV Neutralization.
J.Virol., 97, 2023
|
|
8G9Y
 
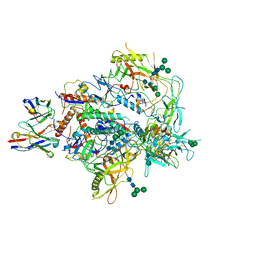 | | Cryo-EM structure of vFP49.02 Fab in complex with HIV-1 Env BG505 DS-SOSIP.664 (conformation 3) | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Envelope glycoprotein gp120, ... | | 著者 | Changela, A, Gorman, J, Kwong, P.D. | | 登録日 | 2023-02-22 | | 公開日 | 2023-04-19 | | 最終更新日 | 2023-06-14 | | 実験手法 | ELECTRON MICROSCOPY (4.28 Å) | | 主引用文献 | Diverse Murine Vaccinations Reveal Distinct Antibody Classes to Target Fusion Peptide and Variation in Peptide Length to Improve HIV Neutralization.
J.Virol., 97, 2023
|
|
7RJF
 
 | | MOPD-1 mutant-L47W | | 分子名称: | MALONATE ION, ZINC ION, [L47W]MOPD-1 | | 著者 | Huawu, Y, Conan, K.W, Gordon, J.K, Brett, M.C, Yen-Hua, H, David, J.C. | | 登録日 | 2021-07-20 | | 公開日 | 2021-10-27 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | Rational Design of Potent Peptide Inhibitors of the PD-1:PD-L1 Interaction for Cancer Immunotherapy.
J.Am.Chem.Soc., 143, 2021
|
|
6OL3
 
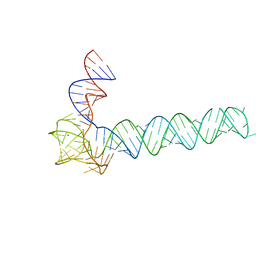 | | Crystal structure of an adenovirus virus-associated RNA | | 分子名称: | Adenovirus Virus-Associated (VA) RNA I apical and central domains, POTASSIUM ION | | 著者 | Hood, I.V, Gordon, J.M, Bou-Nader, C, Henderson, F.V, Bahmanjah, S, Zhang, J. | | 登録日 | 2019-04-15 | | 公開日 | 2019-07-03 | | 最終更新日 | 2024-03-13 | | 実験手法 | X-RAY DIFFRACTION (2.74 Å) | | 主引用文献 | Crystal structure of an adenovirus virus-associated RNA.
Nat Commun, 10, 2019
|
|
7TXD
 
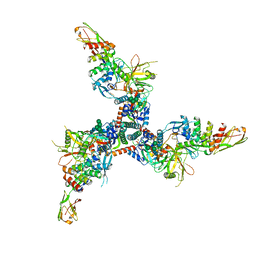 | | Cryo-EM structure of BG505 SOSIP HIV-1 Env trimer in complex with CD4 receptor (D1D2) and broadly neutralizing darpin bnD.9 | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Broadly neutralizing darpin bnd.9, ... | | 著者 | Cerutti, G, Gorman, J, Kwong, P.D, Shapiro, L. | | 登録日 | 2022-02-08 | | 公開日 | 2023-04-12 | | 最終更新日 | 2023-09-27 | | 実験手法 | ELECTRON MICROSCOPY (3.87 Å) | | 主引用文献 | Trapping the HIV-1 V3 loop in a helical conformation enables broad neutralization.
Nat.Struct.Mol.Biol., 30, 2023
|
|
5F89
 
 | |
1LIT
 
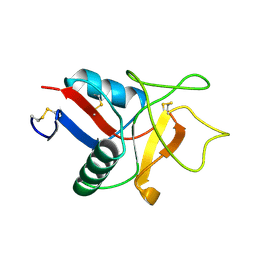 | | HUMAN LITHOSTATHINE | | 分子名称: | LITHOSTATHINE | | 著者 | Bertrand, J.A, Pignol, D, Bernard, J.-P, Verdier, J.-M, Dagorn, J.-C, Fontacilla-Camps, J.C. | | 登録日 | 1996-01-17 | | 公開日 | 1997-01-11 | | 最終更新日 | 2011-07-13 | | 実験手法 | X-RAY DIFFRACTION (1.55 Å) | | 主引用文献 | Crystal structure of human lithostathine, the pancreatic inhibitor of stone formation.
EMBO J., 15, 1996
|
|
7S01
 
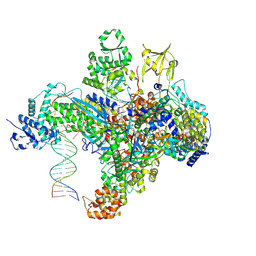 | | X-ray structure of the phage AR9 non-virion RNA polymerase holoenzyme in complex with a forked oligonucleotide containing the P077 promoter | | 分子名称: | DNA-directed RNA polymerase, DNA-directed RNA polymerase beta subunit, DNA-directed RNA polymerase beta' subunit, ... | | 著者 | Leiman, P.G, Sokolova, M.L, Gordeeva, J, Fraser, A, Severinov, K.V. | | 登録日 | 2021-08-28 | | 公開日 | 2022-07-06 | | 最終更新日 | 2024-05-29 | | 実験手法 | X-RAY DIFFRACTION (3.4 Å) | | 主引用文献 | Structural basis of template strand deoxyuridine promoter recognition by a viral RNA polymerase.
Nat Commun, 13, 2022
|
|
8VUE
 
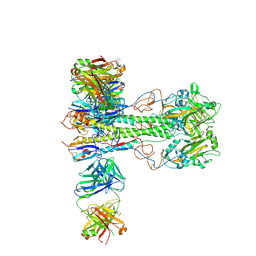 | | L5A7 Fab bound to Indonesia2005 Hemagglutinin | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Hemagglutinin HA1 chain, Hemagglutinin HA2 chain, ... | | 著者 | Olia, A.S, Gorman, J, Kwong, P.D. | | 登録日 | 2024-01-29 | | 公開日 | 2024-05-22 | | 最終更新日 | 2024-06-26 | | 実験手法 | ELECTRON MICROSCOPY (3.59 Å) | | 主引用文献 | Anti-idiotype isolation of a broad and potent influenza A virus-neutralizing human antibody.
Front Immunol, 15, 2024
|
|
1M6O
 
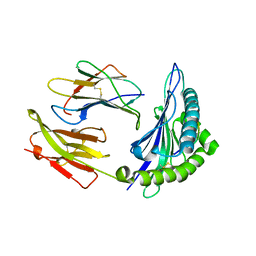 | | Crystal Structure of HLA B*4402 in complex with HLA DPA*0201 peptide | | 分子名称: | Beta-2-microglobulin, HLA DPA*0201 peptide, HLA class I histocompatibility antigen, ... | | 著者 | Macdonald, W.A, Purcell, A.W, Williams, D.S, Mifsud, N.A, Ely, L.K, Gorman, J.J, Clements, C.S, Kjer-Nielsen, L, Koelle, D.M, Brooks, A.G, Lovrecz, G.O, Lu, L, Rossjohn, J, McCluskey, J. | | 登録日 | 2002-07-17 | | 公開日 | 2003-09-02 | | 最終更新日 | 2011-07-13 | | 実験手法 | X-RAY DIFFRACTION (1.6 Å) | | 主引用文献 | A naturally selected dimorphism within the HLA-B44 supertype alters class I structure, peptide repertoire, and T cell recognition.
J.Exp.Med., 198, 2003
|
|
1N2R
 
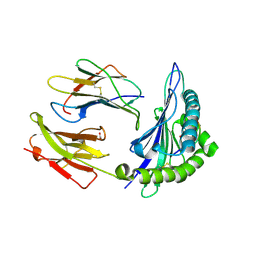 | | A natural selected dimorphism in HLA B*44 alters self, peptide reportoire and T cell recognition. | | 分子名称: | ACETIC ACID, Beta-2-microglobulin, HLA DPA*0201 PEPTIDE, ... | | 著者 | Macdonald, W.A, Purcell, A.W, Williams, D.S, Mifsud, N, Ely, L.K, Gorman, J.J, Clements, C.S, Kjer-Nielsen, L, Koelle, D.M, Brooks, A.G, Lovrecz, G.O, Lu, L, Rossjohn, J, McCluskey, J. | | 登録日 | 2002-10-24 | | 公開日 | 2004-03-16 | | 最終更新日 | 2011-07-13 | | 実験手法 | X-RAY DIFFRACTION (1.7 Å) | | 主引用文献 | A naturally selected dimorphism within the HLA-B44 supertype alters class I structure, peptide repertoire, and T cell recognition.
J.Exp.Med., 198, 2003
|
|
2YMD
 
 | | Crystal structure of a mutant binding protein (5HTBP-AChBP) in complex with serotonin (5-hydroxytryptamine) | | 分子名称: | GLYCEROL, PHOSPHATE ION, SEROTONIN, ... | | 著者 | Kesters, D, Thompson, A.J, Brams, M, Elk, R.v, Spurny, R, Geitmann, M, Villalgordo, J.M, Guskov, A, Danielson, U.H, Lummis, S.C.R, Smit, A.B, Ulens, C. | | 登録日 | 2012-10-09 | | 公開日 | 2012-12-12 | | 最終更新日 | 2023-12-20 | | 実験手法 | X-RAY DIFFRACTION (1.96 Å) | | 主引用文献 | Structural Basis of Ligand Recognition in 5-Ht(3) Receptors.
Embo Rep., 14, 2013
|
|
1U59
 
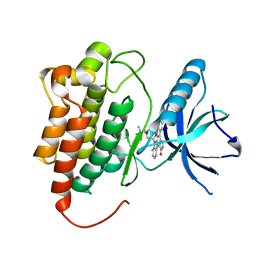 | | Crystal Structure of the ZAP-70 Kinase Domain in Complex with Staurosporine | | 分子名称: | STAUROSPORINE, Tyrosine-protein kinase ZAP-70 | | 著者 | Jin, L, Pluskey, S, Petrella, E.C, Cantin, S.M, Gorga, J.C, Rynkiewicz, M.J, Pandey, P, Strickler, J.E, Babine, R.E, Weaver, D.T, Seidl, K.J. | | 登録日 | 2004-07-27 | | 公開日 | 2004-08-17 | | 最終更新日 | 2023-08-23 | | 実験手法 | X-RAY DIFFRACTION (2.3 Å) | | 主引用文献 | The Three-dimensional Structure of the ZAP-70 Kinase Domain in Complex with Staurosporine: IMPLICATIONS FOR THE DESIGN OF SELECTIVE INHIBITORS
J.Biol.Chem., 279, 2004
|
|
2YME
 
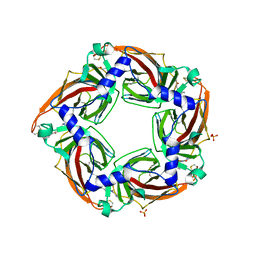 | | Crystal structure of a mutant binding protein (5HTBP-AChBP) in complex with granisetron | | 分子名称: | 1-methyl-N-[(1R,5S)-9-methyl-9-azabicyclo[3.3.1]nonan-3-yl]indazole-3-carboxamide, 2-acetamido-2-deoxy-beta-D-glucopyranose, PHOSPHATE ION, ... | | 著者 | Kesters, D, Thompson, A.J, Brams, M, Elk, R.v, Spurny, R, Geitmann, M, Villalgordo, J.M, Guskov, A, Danielson, U.H, Lummis, S.C.R, Smit, A.B, Ulens, C. | | 登録日 | 2012-10-09 | | 公開日 | 2012-12-26 | | 最終更新日 | 2023-12-20 | | 実験手法 | X-RAY DIFFRACTION (2.4 Å) | | 主引用文献 | Structural Basis of Ligand Recognition in 5-Ht3 Receptors.
Embo Rep., 14, 2013
|
|
7TZ0
 
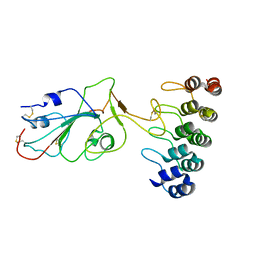 | |
7TYZ
 
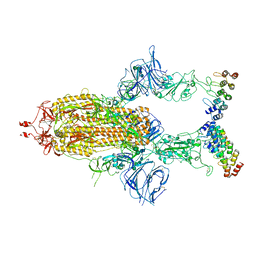 | | Cryo-EM structure of SARS-CoV-2 spike in complex with FSR22, an anti-SARS-CoV-2 DARPin | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, DARPin FSR22, ... | | 著者 | Kwon, Y.D, Gorman, J, Kwong, P.D. | | 登録日 | 2022-02-15 | | 公開日 | 2022-12-07 | | 最終更新日 | 2023-03-15 | | 実験手法 | ELECTRON MICROSCOPY (3.51 Å) | | 主引用文献 | A potent and broad neutralization of SARS-CoV-2 variants of concern by DARPins.
Nat.Chem.Biol., 19, 2023
|
|
8DW2
 
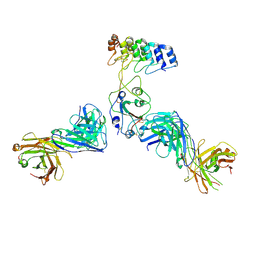 | | Cryo-EM structure of SARS-CoV-2 RBD in complex with anti-SARS-CoV-2 DARPin,SR22, and two antibody Fabs, S309 and CR3022 | | 分子名称: | Antibody CR3022 heavy chain, Antibody CR3022 light chain, Antibody S309 heavy chain, ... | | 著者 | Kwon, Y.D, Gorman, J, Kwong, P.D. | | 登録日 | 2022-07-30 | | 公開日 | 2022-12-07 | | 最終更新日 | 2023-03-15 | | 実験手法 | ELECTRON MICROSCOPY (4.11 Å) | | 主引用文献 | A potent and broad neutralization of SARS-CoV-2 variants of concern by DARPins.
Nat.Chem.Biol., 19, 2023
|
|
8DW3
 
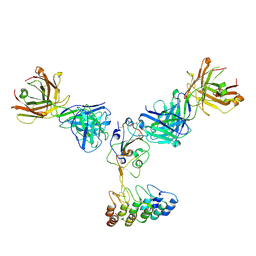 | | Cryo-EM structure of SARS-CoV-2 RBD in complex with anti-SARS-CoV-2 DARPin,SR16m, and two antibody Fabs, S309 and CR3022 | | 分子名称: | Anti-SARS-CoV-2 DARPin SR16m, Antibody S309 light chain, Spike protein S1, ... | | 著者 | Kwon, Y.D, Gorman, J, Kwong, P.D. | | 登録日 | 2022-07-30 | | 公開日 | 2022-12-07 | | 最終更新日 | 2023-03-15 | | 実験手法 | ELECTRON MICROSCOPY (4.26 Å) | | 主引用文献 | A potent and broad neutralization of SARS-CoV-2 variants of concern by DARPins.
Nat.Chem.Biol., 19, 2023
|
|
1XX9
 
 | | Crystal Structure of the FXIa Catalytic Domain in Complex with EcotinM84R | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Coagulation factor XI, Ecotin | | 著者 | Jin, L, Pandey, P, Babine, R.E, Gorga, J.C, Seidl, K.J, Gelfand, E, Weaver, D.T, Abdel-Meguid, S.S, Strickler, J.E. | | 登録日 | 2004-11-04 | | 公開日 | 2004-11-16 | | 最終更新日 | 2023-08-23 | | 実験手法 | X-RAY DIFFRACTION (2.2 Å) | | 主引用文献 | Crystal Structures of the FXIa Catalytic Domain in Complex with Ecotin Mutants Reveal Substrate-like Interactions
J.Biol.Chem., 280, 2005
|
|
1XXD
 
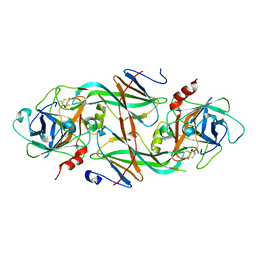 | | Crystal Structure of the FXIa Catalytic Domain in Complex with mutated Ecotin | | 分子名称: | Coagulation factor XI, Ecotin | | 著者 | Jin, L, Pandey, P, Babine, R.E, Gorga, J.C, Seidl, K.J, Gelfand, E, Weaver, D.T, Abdel-Meguid, S.S, Strickler, J.E. | | 登録日 | 2004-11-04 | | 公開日 | 2004-11-16 | | 最終更新日 | 2023-08-23 | | 実験手法 | X-RAY DIFFRACTION (2.91 Å) | | 主引用文献 | Crystal Structures of the FXIa Catalytic Domain in Complex with Ecotin Mutants Reveal Substrate-like Interactions
J.Biol.Chem., 280, 2005
|
|
1XXF
 
 | | Crystal Structure of the FXIa Catalytic Domain in Complex with Ecotin Mutant (EcotinP) | | 分子名称: | Coagulation factor XI, Ecotin, SODIUM ION | | 著者 | Jin, L, Pandey, P, Babine, R.E, Gorga, J.C, Seidl, K.J, Gelfand, E, Weaver, D.T, Abdel-Meguid, S.S, Strickler, J.E. | | 登録日 | 2004-11-04 | | 公開日 | 2004-11-16 | | 最終更新日 | 2023-08-23 | | 実験手法 | X-RAY DIFFRACTION (2.6 Å) | | 主引用文献 | Crystal Structures of the FXIa Catalytic Domain in Complex with Ecotin Mutants Reveal Substrate-like Interactions
J.Biol.Chem., 280, 2005
|
|
3CK9
 
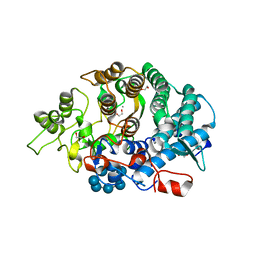 | | B. thetaiotaomicron SusD with maltoheptaose | | 分子名称: | 1,2-ETHANEDIOL, CALCIUM ION, SusD, ... | | 著者 | Koropatkin, N.M, Martens, E.C, Gordon, J.I, Smith, T.J. | | 登録日 | 2008-03-14 | | 公開日 | 2008-05-20 | | 最終更新日 | 2023-08-30 | | 実験手法 | X-RAY DIFFRACTION (2.2 Å) | | 主引用文献 | Starch catabolism by a prominent human gut symbiont is directed by the recognition of amylose helices.
Structure, 16, 2008
|
|
3CK7
 
 | | B. thetaiotaomicron SusD with alpha-cyclodextrin | | 分子名称: | CALCIUM ION, Cyclohexakis-(1-4)-(alpha-D-glucopyranose), SusD | | 著者 | Koropatkin, N.M, Martens, E.C, Gordon, J.I, Smith, T.J. | | 登録日 | 2008-03-14 | | 公開日 | 2008-04-01 | | 最終更新日 | 2023-08-30 | | 実験手法 | X-RAY DIFFRACTION (2.1 Å) | | 主引用文献 | Starch catabolism by a prominent human gut symbiont is directed by the recognition of amylose helices.
Structure, 16, 2008
|
|
