5XF9
 
 | | Crystal structure of NAD+-reducing [NiFe]-hydrogenase in the air-oxidized state | | Descriptor: | CARBONMONOXIDE-(DICYANO) IRON, FE2/S2 (INORGANIC) CLUSTER, FLAVIN MONONUCLEOTIDE, ... | | Authors: | Shomura, Y, Taketa, M, Nakashima, H, Tai, H, Nakagawa, H, Ikeda, Y, Ishii, M, Igarashi, Y, Nishihara, H, Yoon, K.S, Ogo, S, Hirota, S, Higuchi, Y. | | Deposit date: | 2017-04-09 | | Release date: | 2017-08-23 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.58 Å) | | Cite: | Structural basis of the redox switches in the NAD(+)-reducing soluble [NiFe]-hydrogenase
Science, 357, 2017
|
|
1HJP
 
 | | HOLLIDAY JUNCTION BINDING PROTEIN RUVA FROM E. COLI | | Descriptor: | RUVA | | Authors: | Nishino, T, Ariyoshi, M, Iwasaki, H, Shinagawa, H, Morikawa, K. | | Deposit date: | 1997-08-21 | | Release date: | 1998-02-25 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Functional Analyses of the Domain Structure in the Holliday Junction Binding Protein Ruva
Structure, 6, 1998
|
|
2LI5
 
 | | NMR structure of Atg8-Atg7C30 complex | | Descriptor: | Autophagy-related protein 8, Ubiquitin-like modifier-activating enzyme ATG7 | | Authors: | Kumeta, H, Satoo, K, Noda, N.N, Fujioka, Y, Ogura, K, Nakatogawa, H, Ohsumi, Y, Inagaki, F. | | Deposit date: | 2011-08-23 | | Release date: | 2011-11-16 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Structural basis of Atg8 activation by a homodimeric E1, Atg7.
Mol.Cell, 44, 2011
|
|
1ON8
 
 | | Crystal structure of mouse alpha-1,4-N-acetylhexosaminyltransferase (EXTL2) with UDP and GlcUAb(1-3)Galb(1-O)-naphthalenelmethanol an acceptor substrate analog | | Descriptor: | 1,2-ETHANEDIOL, Alpha-1,4-N-acetylhexosaminyltransferase EXTL2, MANGANESE (II) ION, ... | | Authors: | Pedersen, L.C, Dong, J, Taniguchi, F, Kitagawa, H, Krahn, J.M, Pedersen, L.G, Sugahara, K, Negishi, M. | | Deposit date: | 2003-02-27 | | Release date: | 2003-04-22 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Crystal structure of an alpha-1,4-N-acetylhexosaminyltransferase (EXTL2), a member of the exostosin gene family involved in heparan sulfate biosynthesis
J.Biol.Chem., 278, 2003
|
|
1OMZ
 
 | | crystal structure of mouse alpha-1,4-N-acetylhexosaminyltransferase (EXTL2) in complex with UDPGalNAc | | Descriptor: | 1,2-ETHANEDIOL, Alpha-1,4-N-acetylhexosaminyltransferase EXTL2, MANGANESE (II) ION, ... | | Authors: | Pedersen, L.C, Dong, J, Taniguchi, F, Kitagawa, H, Krahn, J.M, Pedersen, L.G, Sugahara, K, Negishi, M. | | Deposit date: | 2003-02-26 | | Release date: | 2003-04-22 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structure of an alpha-1,4-N-acetylhexosaminyltransferase (EXTL2),
a member of the exostosin gene family involved in heparan sulfate biosynthesis
J.Biol.Chem., 278, 2003
|
|
1FGG
 
 | | CRYSTAL STRUCTURE OF 1,3-GLUCURONYLTRANSFERASE I (GLCAT-I) COMPLEXED WITH GAL-GAL-XYL, UDP, AND MN2+ | | Descriptor: | GLUCURONYLTRANSFERASE I, MANGANESE (II) ION, UNKNOWN ATOM OR ION, ... | | Authors: | Pedersen, L.C, Tsuchida, K, Kitagawa, H, Sugahara, K, Darden, T.A. | | Deposit date: | 2000-07-28 | | Release date: | 2001-01-31 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Heparan/chondroitin sulfate biosynthesis. Structure and mechanism of human glucuronyltransferase I.
J.Biol.Chem., 275, 2000
|
|
1ON6
 
 | | Crystal structure of mouse alpha-1,4-N-acetylhexosaminotransferase (EXTL2) in complex with UDPGlcNAc | | Descriptor: | 1,2-ETHANEDIOL, Alpha-1,4-N-acetylhexosaminyltransferase EXTL2, MANGANESE (II) ION, ... | | Authors: | Pedersen, L.C, Dong, J, Taniguchi, F, Kitagawa, H, Krahn, J.M, Pedersen, L.G, Sugahara, K, Negishi, M. | | Deposit date: | 2003-02-27 | | Release date: | 2003-04-22 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal Structure of an alpha-1,4-N-acetylhexosaminyltransferase (EXTL2), a member of the exostosin gene family involved in heparan sulfate biosynthesis
J.Biol.Chem., 278, 2003
|
|
7D92
 
 | | Crystal Structure of the Na+,K+-ATPase in the E2P state with bound Mg2+ and anthroylouabain (P4(3)2(1)2 symmetry) | | Descriptor: | 1,2-DIOLEOYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Kanai, R, Cornelius, F, Ogawa, H, Motoyama, K, Vilsen, B, Toyoshima, C. | | Deposit date: | 2020-10-12 | | Release date: | 2021-01-27 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (3.9 Å) | | Cite: | Binding of cardiotonic steroids to Na + ,K + -ATPase in the E2P state.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
7D93
 
 | | Crystal Structure of the Na+,K+-ATPase in the E2P state with bound Mg2+ and anthroylouabain (P2(1)2(1)2(1) symmetry) | | Descriptor: | 1,2-DIOLEOYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Kanai, R, Cornelius, F, Ogawa, H, Motoyama, K, Vilsen, B, Toyoshima, C. | | Deposit date: | 2020-10-12 | | Release date: | 2021-01-27 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (3.65 Å) | | Cite: | Binding of cardiotonic steroids to Na + ,K + -ATPase in the E2P state.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
2RUH
 
 | | Chemical Shift Assignments for MIP and MDM2 in bound state | | Descriptor: | E3 ubiquitin-protein ligase Mdm2 | | Authors: | Nagata, T, Shirakawa, K, Kobayashi, N, Shiheido, H, Horisawa, K, Katahira, M, Doi, N, Yanagawa, H. | | Deposit date: | 2014-06-03 | | Release date: | 2014-10-15 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Structural Basis for Inhibition of the MDM2:p53 Interaction by an Optimized MDM2-Binding Peptide Selected with mRNA Display
Plos One, 9, 2014
|
|
7CDC
 
 | | Crystal structure of LSD1-CoREST in complex with PRSFLVRRP peptide | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, GLYCEROL, Lysine-specific histone demethylase 1A, ... | | Authors: | Kikuchi, M, Kitagawa, H, Sato, S, Umezawa, N, Higuchi, T, Umehara, T. | | Deposit date: | 2020-06-19 | | Release date: | 2021-06-23 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.64 Å) | | Cite: | Structure-Based Identification of Potent Lysine-Specific Demethylase 1 Inhibitor Peptides and Temporary Cyclization to Enhance Proteolytic Stability and Cell Growth-Inhibitory Activity.
J.Med.Chem., 64, 2021
|
|
7CDF
 
 | | Crystal structure of LSD1-CoREST in complex with PRSFLVRRK peptide | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, GLYCEROL, Lysine-specific histone demethylase 1A, ... | | Authors: | Kikuchi, M, Kitagawa, H, Sato, S, Umezawa, N, Higuchi, T, Umehara, T. | | Deposit date: | 2020-06-19 | | Release date: | 2021-06-23 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.68 Å) | | Cite: | Structure-Based Identification of Potent Lysine-Specific Demethylase 1 Inhibitor Peptides and Temporary Cyclization to Enhance Proteolytic Stability and Cell Growth-Inhibitory Activity.
J.Med.Chem., 64, 2021
|
|
7CDD
 
 | | Crystal structure of LSD1-CoREST in complex with PRSFLVRR peptide | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, GLYCEROL, Lysine-specific histone demethylase 1A, ... | | Authors: | Kikuchi, M, Kitagawa, H, Sato, S, Umezawa, N, Higuchi, T, Umehara, T. | | Deposit date: | 2020-06-19 | | Release date: | 2021-06-23 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.76 Å) | | Cite: | Structure-Based Identification of Potent Lysine-Specific Demethylase 1 Inhibitor Peptides and Temporary Cyclization to Enhance Proteolytic Stability and Cell Growth-Inhibitory Activity.
J.Med.Chem., 64, 2021
|
|
7CDG
 
 | | Crystal structure of LSD1-CoREST in complex with PRSFLVRRR peptide | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, GLYCEROL, Lysine-specific histone demethylase 1A, ... | | Authors: | Kikuchi, M, Kitagawa, H, Sato, S, Umezawa, N, Higuchi, T, Umehara, T. | | Deposit date: | 2020-06-19 | | Release date: | 2021-06-23 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structure-Based Identification of Potent Lysine-Specific Demethylase 1 Inhibitor Peptides and Temporary Cyclization to Enhance Proteolytic Stability and Cell Growth-Inhibitory Activity.
J.Med.Chem., 64, 2021
|
|
7CDE
 
 | | Crystal structure of LSD1-CoREST in complex with PRSFLVRKR peptide | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, GLYCEROL, Lysine-specific histone demethylase 1A, ... | | Authors: | Kikuchi, M, Kitagawa, H, Sato, S, Umezawa, N, Higuchi, T, Umehara, T. | | Deposit date: | 2020-06-19 | | Release date: | 2021-06-23 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.68 Å) | | Cite: | Structure-Based Identification of Potent Lysine-Specific Demethylase 1 Inhibitor Peptides and Temporary Cyclization to Enhance Proteolytic Stability and Cell Growth-Inhibitory Activity.
J.Med.Chem., 64, 2021
|
|
1OMX
 
 | | Crystal structure of mouse alpha-1,4-N-acetylhexosaminyltransferase (EXTL2) | | Descriptor: | 1,2-ETHANEDIOL, Alpha-1,4-N-acetylhexosaminyltransferase EXTL2 | | Authors: | Pedersen, L.C, Dong, J, Taniguchi, F, Kitagawa, H, Krahn, J.M, Pedersen, L.G, Sugahara, K, Negishi, M. | | Deposit date: | 2003-02-26 | | Release date: | 2003-04-22 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal structure of an alpha-1,4-N-acetylhexosaminyltransferase (EXTL2), a member of the exostosin gene family involved in heparan sulfate biosynthesis
J.Biol.Chem., 278, 2003
|
|
5ZCN
 
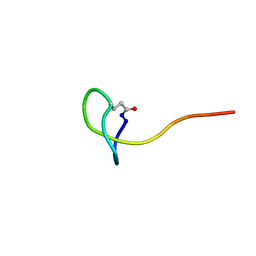 | | Solution NMR structure of a new lasso peptide brevunsin | | Descriptor: | brevunsin | | Authors: | Hemmi, H, Kodani, S, Miyake, Y, Kaweewan, I, Nakagawa, H. | | Deposit date: | 2018-02-19 | | Release date: | 2018-10-17 | | Last modified: | 2024-10-09 | | Method: | SOLUTION NMR | | Cite: | Heterologous production of a new lasso peptide brevunsin in Sphingomonas subterranea
J. Ind. Microbiol. Biotechnol., 45, 2018
|
|
7D94
 
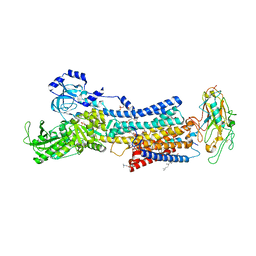 | | Crystal Structure of the Na+,K+-ATPase in the E2P state with bound one Mg2+ and one Rb+ in the presence of bufalin | | Descriptor: | 1,2-DIOLEOYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Kanai, R, Cornelius, F, Ogawa, H, Motoyama, K, Vilsen, B, Toyoshima, C. | | Deposit date: | 2020-10-12 | | Release date: | 2021-01-27 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Binding of cardiotonic steroids to Na + ,K + -ATPase in the E2P state.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
7D91
 
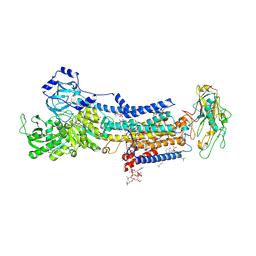 | | Crystal Structure of the Na+,K+-ATPase in the E2P state with bound Mg2+ (P4(3)2(1)2 symmetry) | | Descriptor: | 1,2-DIOLEOYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Kanai, R, Cornelius, F, Ogawa, H, Motoyama, K, Vilsen, B, Toyoshima, C. | | Deposit date: | 2020-10-12 | | Release date: | 2021-01-27 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (3.35 Å) | | Cite: | Binding of cardiotonic steroids to Na + ,K + -ATPase in the E2P state.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
1VCL
 
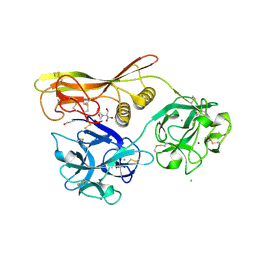 | | Crystal Structure of Hemolytic Lectin CEL-III | | Descriptor: | 2-[BIS-(2-HYDROXY-ETHYL)-AMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, CALCIUM ION, CHLORIDE ION, ... | | Authors: | Uchida, T, Yamasaki, T, Eto, S, Sugawara, H, Kurisu, G, Nakagawa, A, Kusunoki, M, Hatakeyama, T. | | Deposit date: | 2004-03-09 | | Release date: | 2004-09-07 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal Structure of the Hemolytic Lectin CEL-III Isolated from the Marine Invertebrate Cucumaria echinata: IMPLICATIONS OF DOMAIN STRUCTURE FOR ITS MEMBRANE PORE-FORMATION MECHANISM
J.Biol.Chem., 279, 2004
|
|
3VW6
 
 | | Crystal structure of human apoptosis signal-regulating kinase 1 (ASK1) with imidazopyridine inhibitor | | Descriptor: | 4-tert-butyl-N-[6-(1H-imidazol-1-yl)imidazo[1,2-a]pyridin-2-yl]benzamide, Mitogen-activated protein kinase kinase kinase 5 | | Authors: | Terao, Y, Suzuki, H, Yoshikawa, M, Yashiro, H, Takekawa, S, Fujitani, Y, Okada, K, Inoue, Y, Yamamoto, Y, Nakagawa, H, Yao, S, Kawamoto, T, Uchikawa, O. | | Deposit date: | 2012-08-06 | | Release date: | 2012-10-31 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Design and biological evaluation of imidazo[1,2-a]pyridines as novel and potent ASK1 inhibitors.
Bioorg. Med. Chem. Lett., 22, 2012
|
|
2SIC
 
 | | REFINED CRYSTAL STRUCTURE OF THE COMPLEX OF SUBTILISIN BPN' AND STREPTOMYCES SUBTILISIN INHIBITOR AT 1.8 ANGSTROMS RESOLUTION | | Descriptor: | CALCIUM ION, STREPTOMYCES SUBTILISIN INHIBITOR (SSI), SUBTILISIN BPN' | | Authors: | Mitsui, Y, Takeuchi, Y, Hirono, S, Akagawa, H, Nakamura, K.T. | | Deposit date: | 1991-04-01 | | Release date: | 1993-04-15 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Refined crystal structure of the complex of subtilisin BPN' and Streptomyces subtilisin inhibitor at 1.8 A resolution.
J.Mol.Biol., 221, 1991
|
|
3W5B
 
 | | Crystal structure of the recombinant SERCA1a (calcium pump of fast twitch skeletal muscle) in the E1.Mg2+ state | | Descriptor: | 2',3'-O-[(1r)-2,4,6-trinitrocyclohexa-2,5-diene-1,1-diyl]adenosine 5'-(dihydrogen phosphate), MAGNESIUM ION, PHOSPHATIDYLETHANOLAMINE, ... | | Authors: | Toyoshima, C, Iwasawa, S, Ogawa, H, Hirata, A, Tsueda, J, Inesi, G. | | Deposit date: | 2013-01-27 | | Release date: | 2013-03-06 | | Last modified: | 2013-03-27 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Crystal structures of the calcium pump and sarcolipin in the Mg2+-bound E1 state.
Nature, 495, 2013
|
|
3W5C
 
 | | Crystal structure of the calcium pump in the E2 state free from exogenous inhibitors | | Descriptor: | PHOSPHATIDYLETHANOLAMINE, SERCA1a, SODIUM ION | | Authors: | Toyoshima, C, Iwasawa, S, Ogawa, H, Hirata, A, Tsueda, J, Inesi, G. | | Deposit date: | 2013-01-27 | | Release date: | 2013-03-06 | | Last modified: | 2013-03-27 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structures of the calcium pump and sarcolipin in the Mg2+-bound E1 state.
Nature, 495, 2013
|
|
3WGV
 
 | | Crystal structure of a Na+-bound Na+,K+-ATPase preceding the E1P state with oligomycin | | Descriptor: | 1,2-DIACYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 2-acetamido-2-deoxy-beta-D-glucopyranose, ADENOSINE-5'-DIPHOSPHATE, ... | | Authors: | Kanai, R, Ogawa, H, Vilsen, B, Cornelius, F, Toyoshima, C. | | Deposit date: | 2013-08-09 | | Release date: | 2013-10-09 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal structure of a Na1-bound Na1,K1-ATPase preceding the E1P state
Nature, 502, 2013
|
|
