7N2R
 
 | | AS4.3-PRPF3-HLA*B27 | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, AS4.3 T cell receptor alpha chain, ... | | Authors: | Yang, X, Jude, K.M, Garcia, K.C. | | Deposit date: | 2021-05-29 | | Release date: | 2022-12-07 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.28 Å) | | Cite: | Autoimmunity-associated T cell receptors recognize HLA-B*27-bound peptides.
Nature, 612, 2022
|
|
7N2N
 
 | | TCR-antigen complex AS4.2-PRPF3-HLA*B27 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Beta-2-microglobulin, GLYCEROL, ... | | Authors: | Yang, X, Jude, K.M, Garcia, K.C. | | Deposit date: | 2021-05-29 | | Release date: | 2022-12-07 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Autoimmunity-associated T cell receptors recognize HLA-B*27-bound peptides.
Nature, 612, 2022
|
|
4XL1
 
 | | Complex of Notch1 (EGF11-13) bound to Delta-like 4 (N-EGF1) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, ... | | Authors: | Luca, V.C, Jude, K.M, Garcia, K.C. | | Deposit date: | 2015-01-13 | | Release date: | 2015-03-04 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural biology. Structural basis for Notch1 engagement of Delta-like 4.
Science, 347, 2015
|
|
4XLW
 
 | | Complex of Notch1 (EGF11-13) bound to Delta-like 4 (N-EGF2) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, Delta-like protein, ... | | Authors: | Luca, V.C, Jude, K.M, Garcia, K.C. | | Deposit date: | 2015-01-13 | | Release date: | 2015-03-04 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (3.39 Å) | | Cite: | Structural biology. Structural basis for Notch1 engagement of Delta-like 4.
Science, 347, 2015
|
|
4XT3
 
 | | Structure of a viral GPCR bound to human chemokine CX3CL1 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Fractalkine, G-protein coupled receptor homolog US28, ... | | Authors: | Burg, J.S, Jude, K.M, Waghray, D, Garcia, K.C. | | Deposit date: | 2015-01-22 | | Release date: | 2015-03-04 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (3.801 Å) | | Cite: | Structural biology. Structural basis for chemokine recognition and activation of a viral G protein-coupled receptor.
Science, 347, 2015
|
|
4XT1
 
 | | Structure of a nanobody-bound viral GPCR bound to human chemokine CX3CL1 | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, CHOLESTEROL, Fractalkine, ... | | Authors: | Burg, J.S, Jude, K.M, Waghray, D, Garcia, K.C. | | Deposit date: | 2015-01-22 | | Release date: | 2015-03-04 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.886 Å) | | Cite: | Structural biology. Structural basis for chemokine recognition and activation of a viral G protein-coupled receptor.
Science, 347, 2015
|
|
7S2R
 
 | | nanobody bound to IL-2Rg | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Cytokine receptor common subunit gamma, ... | | Authors: | Glassman, C.R, Jude, K.M, Yen, M, Garcia, K.C. | | Deposit date: | 2021-09-03 | | Release date: | 2022-03-30 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.49 Å) | | Cite: | Facile discovery of surrogate cytokine agonists.
Cell, 185, 2022
|
|
7S2S
 
 | | nanobody bound to Interleukin-2Rbeta | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, IL2Rb-binding nanobody, Interleukin-2 receptor subunit beta, ... | | Authors: | Glassman, C.R, Jude, K.M, Yen, M, Garcia, K.C. | | Deposit date: | 2021-09-03 | | Release date: | 2022-03-30 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.93 Å) | | Cite: | Facile discovery of surrogate cytokine agonists.
Cell, 185, 2022
|
|
2Z31
 
 | | Crystal structure of immune receptor complex | | Descriptor: | H-2 class II histocompatibility antigen, A-U alpha chain, A-U beta chain precursor, ... | | Authors: | Feng, D, Bond, C.J, Ely, L.K, Garcia, K.C. | | Deposit date: | 2007-05-30 | | Release date: | 2007-10-09 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structural evidence for a germline-encoded T cell receptor-major histocompatibility complex interaction 'codon'.
Nat.Immunol., 8, 2007
|
|
3IJ2
 
 | | Ligand-receptor structure | | Descriptor: | Beta-nerve growth factor, Nerve growth factor receptor (TNFR superfamily, member 16) | | Authors: | Feng, D, Garcia, K.C. | | Deposit date: | 2009-08-03 | | Release date: | 2010-01-12 | | Last modified: | 2021-10-13 | | Method: | X-RAY DIFFRACTION (3.75 Å) | | Cite: | Molecular and structural insight into proNGF engagement of p75NTR and sortilin.
J.Mol.Biol., 396, 2010
|
|
4YUE
 
 | | Mouse IL-2 Bound to S4B6 Fab Fragment | | Descriptor: | GLYCEROL, Interleukin-2, S4B6 Fab heavy chain, ... | | Authors: | Spangler, J.B, Jude, K.M, Garcia, K.C. | | Deposit date: | 2015-03-18 | | Release date: | 2015-07-01 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.194 Å) | | Cite: | Antibodies to Interleukin-2 Elicit Selective T Cell Subset Potentiation through Distinct Conformational Mechanisms.
Immunity, 42, 2015
|
|
3GCD
 
 | | Structure of the V. cholerae RTX cysteine protease domain in complex with an aza-Leucine peptide inhibitor | | Descriptor: | INOSITOL HEXAKISPHOSPHATE, RTX toxin RtxA, SODIUM ION, ... | | Authors: | Lupardus, P.J, Garcia, K.C, Shen, A, Bogyo, M. | | Deposit date: | 2009-02-21 | | Release date: | 2009-05-26 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Mechanistic and structural insights into the proteolytic activation of Vibrio cholerae MARTX toxin.
Nat.Chem.Biol., 5, 2009
|
|
4YQX
 
 | | Mouse IL-2 Bound to JES6-1 scFv Fragment | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Interleukin-2, JES6-1 VH domain, ... | | Authors: | Spangler, J.B, Luca, V.C, Jude, K.M, Garcia, K.C. | | Deposit date: | 2015-03-13 | | Release date: | 2015-07-01 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.826 Å) | | Cite: | Antibodies to Interleukin-2 Elicit Selective T Cell Subset Potentiation through Distinct Conformational Mechanisms.
Immunity, 42, 2015
|
|
3JVF
 
 | | Crystal structure of an Interleukin-17 receptor complex | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, ... | | Authors: | Ely, L.K, Garcia, K.C. | | Deposit date: | 2009-09-16 | | Release date: | 2009-10-20 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Structural basis of receptor sharing by interleukin 17 cytokines.
Nat.Immunol., 10, 2009
|
|
2Z35
 
 | | Crystal structure of immune receptor | | Descriptor: | T-cell receptor alpha-chain, T-cell receptor beta-chain | | Authors: | Feng, D, Bond, C.J, Ely, L.K, Garcia, K.C. | | Deposit date: | 2007-06-01 | | Release date: | 2007-10-09 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural evidence for a germline-encoded T cell receptor-major histocompatibility complex interaction 'codon'
Nat.Immunol., 8, 2007
|
|
6BJ3
 
 | | TCR55 in complex with HIV(Pol448-456)/HLA-B35 | | Descriptor: | 1,2-ETHANEDIOL, Beta-2-microglobulin, HIV Pol B35 peptide, ... | | Authors: | Jude, K.M, Sibener, L.V, Garcia, K.C. | | Deposit date: | 2017-11-03 | | Release date: | 2018-07-25 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.898 Å) | | Cite: | Isolation of a Structural Mechanism for Uncoupling T Cell Receptor Signaling from Peptide-MHC Binding.
Cell, 174, 2018
|
|
6BJ8
 
 | | TCR55 in complex with Pep20/HLA-B35 | | Descriptor: | Beta-2-microglobulin, GLYCEROL, HLA class I histocompatibility antigen, ... | | Authors: | Jude, K.M, Sibener, L.V, Yang, X, Garcia, K.C. | | Deposit date: | 2017-11-05 | | Release date: | 2018-07-25 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Isolation of a Structural Mechanism for Uncoupling T Cell Receptor Signaling from Peptide-MHC Binding.
Cell, 174, 2018
|
|
3E3Q
 
 | |
3EEB
 
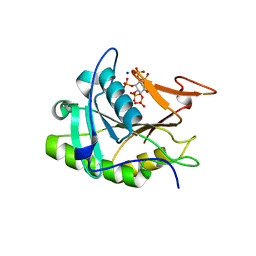 | | Structure of the V. cholerae RTX cysteine protease domain | | Descriptor: | INOSITOL HEXAKISPHOSPHATE, RTX toxin RtxA, SODIUM ION | | Authors: | Lupardus, P.J, Shen, A, Bogyo, M, Garcia, K.C. | | Deposit date: | 2008-09-04 | | Release date: | 2008-10-21 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Small molecule-induced allosteric activation of the Vibrio cholerae RTX cysteine protease domain
Science, 322, 2008
|
|
6BJ2
 
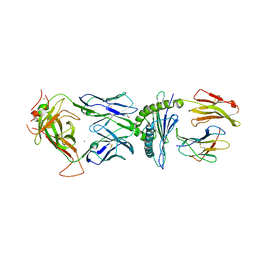 | | TCR589 in complex with HIV(Pol448-456)/HLA-B35 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Beta-2-microglobulin, ... | | Authors: | Jude, K.M, Sibener, L.V, Garcia, K.C. | | Deposit date: | 2017-11-03 | | Release date: | 2018-07-25 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (3.35 Å) | | Cite: | Isolation of a Structural Mechanism for Uncoupling T Cell Receptor Signaling from Peptide-MHC Binding.
Cell, 174, 2018
|
|
6DG6
 
 | | Structure of a de novo designed Interleukin-2/Interleukin-15 mimetic | | Descriptor: | Neoleukin-2/15 | | Authors: | Jude, K.M, Silva, D.-A, Yu, S, Baker, D, Garcia, K.C. | | Deposit date: | 2018-05-16 | | Release date: | 2019-01-16 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.999 Å) | | Cite: | De novo design of potent and selective mimics of IL-2 and IL-15.
Nature, 565, 2019
|
|
2B5I
 
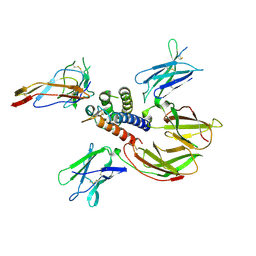 | | cytokine receptor complex | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Cytokine receptor common gamma chain, ... | | Authors: | Wang, X, Rickert, M, Garcia, K.C. | | Deposit date: | 2005-09-28 | | Release date: | 2005-11-29 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structure of the quaternary complex of interleukin-2 with its alpha, beta, and gammac receptors.
Science, 310, 2005
|
|
6E3K
 
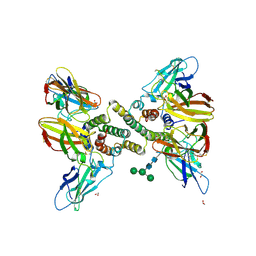 | | Interferon gamma signalling complex with IFNGR1 and IFNGR2 | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Jude, K.M, Mendoza, J.L, Garcia, K.C. | | Deposit date: | 2018-07-14 | | Release date: | 2019-02-27 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (3.25 Å) | | Cite: | Structure of the IFN gamma receptor complex guides design of biased agonists.
Nature, 567, 2019
|
|
6E3L
 
 | | Interferon gamma signalling complex with IFNGR1 and IFNGR2 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CYSTEINE, ... | | Authors: | Jude, K.M, Mendoza, J.L, Garcia, K.C. | | Deposit date: | 2018-07-14 | | Release date: | 2019-02-27 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (3.8 Å) | | Cite: | Structure of the IFN gamma receptor complex guides design of biased agonists.
Nature, 567, 2019
|
|
3E2H
 
 | |
