1NQY
 
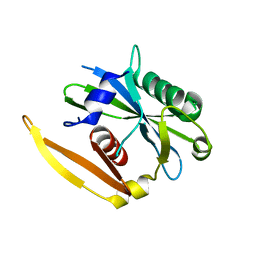 | | The structure of a CoA pyrophosphatase from D. Radiodurans | | Descriptor: | CoA pyrophosphatase (MutT/nudix family protein) | | Authors: | Kang, L.W, Gabelli, S.B, Bianchet, M.A, Xu, W.L, Bessman, M.J, Amzel, L.M. | | Deposit date: | 2003-01-23 | | Release date: | 2003-05-13 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.09 Å) | | Cite: | Structure of a coenzyme A pyrophosphatase from Deinococcus radiodurans: a member of the Nudix family.
J.Bacteriol., 185, 2003
|
|
4K10
 
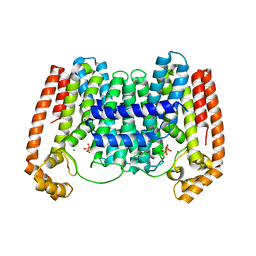 | | Crystal Structure of Leshmaniasis major Farnesyl diphosphate synthase in complex with 3-FLUORO-1-(2-HYDROXY-2,2-DIPHOSPHONOETHYL)PYRIDINIUM and Mg2+ | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, 3-FLUORO-1-(2-HYDROXY-2,2-DIPHOSPHONOETHYL)PYRIDINIUM, ACETATE ION, ... | | Authors: | Aripirala, S, Gabelli, S, Amzel, L.M. | | Deposit date: | 2013-04-04 | | Release date: | 2014-04-02 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural and thermodynamic basis of the inhibition of Leishmania major farnesyl diphosphate synthase by nitrogen-containing bisphosphonates.
Acta Crystallogr.,Sect.D, 70, 2014
|
|
4JZX
 
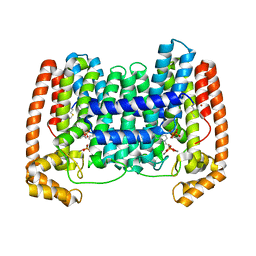 | | Crystal Structure of Leshmaniasis major Farnesyl diphosphate synthase in complex with 3-BUTYL-1-(2,2-DIPHOSPHONOETHYL)PYRIDINIUM, IPP and Ca2+ | | Descriptor: | 3-METHYLBUT-3-ENYL TRIHYDROGEN DIPHOSPHATE, 3-butyl-1-(2,2-diphosphonoethyl)pyridinium, CALCIUM ION, ... | | Authors: | Aripirala, S, Gabelli, S, Amzel, L.M. | | Deposit date: | 2013-04-03 | | Release date: | 2014-03-05 | | Last modified: | 2017-11-15 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural and thermodynamic basis of the inhibition of Leishmania major farnesyl diphosphate synthase by nitrogen-containing bisphosphonates.
Acta Crystallogr.,Sect.D, 70, 2014
|
|
6NCT
 
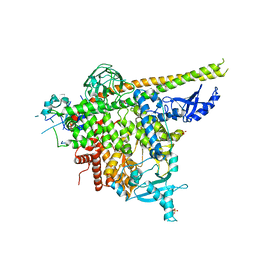 | | Structure of p110alpha/niSH2 - vector data collection | | Descriptor: | Phosphatidylinositol 3-kinase regulatory subunit alpha, Phosphatidylinositol 4,5-bisphosphate 3-kinase catalytic subunit alpha isoform, SULFATE ION, ... | | Authors: | Miller, M.S, Maheshwari, S, Amzel, L.M, Gabelli, S.B. | | Deposit date: | 2018-12-12 | | Release date: | 2019-02-06 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (3.35 Å) | | Cite: | Getting the Most Out of Your Crystals: Data Collection at the New High-Flux, Microfocus MX Beamlines at NSLS-II.
Molecules, 24, 2019
|
|
6NCK
 
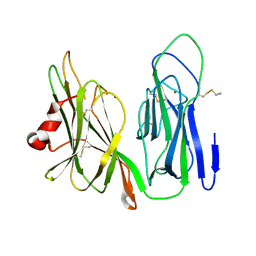 | | Crystal structure of H108A peptidylglycine alpha-hydroxylating monooxygenase (PHM) | | Descriptor: | COPPER (II) ION, NICKEL (II) ION, Peptidyl-glycine alpha-amidating monooxygenase | | Authors: | Miller, M.S, Maheshwari, S, Gabelli, S.B. | | Deposit date: | 2018-12-11 | | Release date: | 2019-02-06 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Getting the Most Out of Your Crystals: Data Collection at the New High-Flux, Microfocus MX Beamlines at NSLS-II.
Molecules, 24, 2019
|
|
6NCI
 
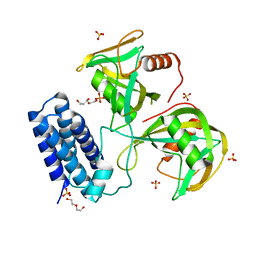 | | Crystal structure of CDP-Chase: Vector data collection | | Descriptor: | D-ribose, DI(HYDROXYETHYL)ETHER, PHOSPHATE ION, ... | | Authors: | Miller, M.S, Shi, W, Gabelli, S.B. | | Deposit date: | 2018-12-11 | | Release date: | 2019-02-06 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.08 Å) | | Cite: | Getting the Most Out of Your Crystals: Data Collection at the New High-Flux, Microfocus MX Beamlines at NSLS-II.
Molecules, 24, 2019
|
|
6NCH
 
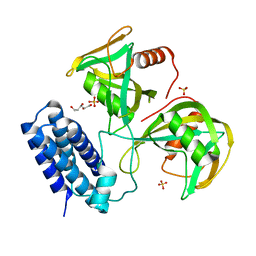 | | Crystal structure of CDP-Chase: Raster data collection | | Descriptor: | D-ribose, PHOSPHATE ION, Phosphohydrolase (MutT/nudix family protein), ... | | Authors: | Miller, M.S, Shi, W, Gabelli, S.B. | | Deposit date: | 2018-12-11 | | Release date: | 2019-02-06 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Getting the Most Out of Your Crystals: Data Collection at the New High-Flux, Microfocus MX Beamlines at NSLS-II.
Molecules, 24, 2019
|
|
6O9C
 
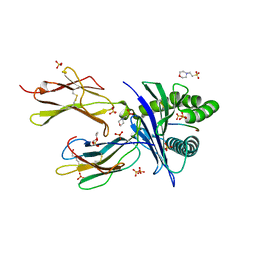 | |
6O9B
 
 | |
8E0P
 
 | | Crystal structure of mouse APCDD1 in fusion with engineered MBP | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 3-[BENZYL(DIMETHYL)AMMONIO]PROPANE-1-SULFONATE, CHLORIDE ION, ... | | Authors: | Hsieh, F.L, Chang, T.H, Gabelli, S.B, Nathans, J. | | Deposit date: | 2022-08-09 | | Release date: | 2023-05-03 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.33 Å) | | Cite: | Structure of WNT inhibitor adenomatosis polyposis coli down-regulated 1 (APCDD1), a cell-surface lipid-binding protein.
Proc.Natl.Acad.Sci.USA, 120, 2023
|
|
7STF
 
 | | Structure of KRAS G12V/HLA-A*03:01 in complex with antibody fragment V2 | | Descriptor: | Beta-2-microglobulin, HLA class I histocompatibility antigen, A alpha chain, ... | | Authors: | Wright, K.M, Gabelli, S.B, Miller, M. | | Deposit date: | 2021-11-12 | | Release date: | 2023-05-31 | | Last modified: | 2023-10-25 | | Method: | ELECTRON MICROSCOPY (3.14 Å) | | Cite: | Hydrophobic interactions dominate the recognition of a KRAS G12V neoantigen.
Nat Commun, 14, 2023
|
|
6NPZ
 
 | | Crystal structure of Akt1 (aa 123-480) kinase with a bisubstrate | | Descriptor: | GLYCEROL, MANGANESE (II) ION, RAC-alpha serine/threonine-protein kinase, ... | | Authors: | Chu, N, Cole, P.A, Gabelli, S.B. | | Deposit date: | 2019-01-18 | | Release date: | 2019-01-30 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.12 Å) | | Cite: | Akt Kinase Activation Mechanisms Revealed Using Protein Semisynthesis.
Cell, 174, 2018
|
|
6UJ8
 
 | | Crystal structure of HLA-B*07:02 with wild-type IDH2 peptide | | Descriptor: | Beta-2-microglobulin, DI(HYDROXYETHYL)ETHER, HLA class I histocompatibility antigen, ... | | Authors: | Miller, M.S, Thirawatananond, P, Gabelli, S.B. | | Deposit date: | 2019-10-02 | | Release date: | 2021-05-05 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Structural engineering of chimeric antigen receptors targeting HLA-restricted neoantigens.
Nat Commun, 12, 2021
|
|
6UJ9
 
 | | Crystal structure of HLA-B*07:02 with R140Q mutant IDH2 peptide in complex with Fab | | Descriptor: | 2-[N-CYCLOHEXYLAMINO]ETHANE SULFONIC ACID, Beta-2-microglobulin, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Miller, M.S, Thirawatananond, P, Aytenfisu, T.Y, Wright, K, Gabelli, S.B. | | Deposit date: | 2019-10-02 | | Release date: | 2021-05-05 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structural engineering of chimeric antigen receptors targeting HLA-restricted neoantigens.
Nat Commun, 12, 2021
|
|
6UJ7
 
 | | Crystal structure of HLA-B*07:02 with R140Q mutant IDH2 peptide | | Descriptor: | Beta-2-microglobulin, HLA class I histocompatibility antigen, B-7 alpha chain, ... | | Authors: | Miller, M.S, Thirawatananond, P, Gabelli, S.B. | | Deposit date: | 2019-10-02 | | Release date: | 2021-05-05 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural engineering of chimeric antigen receptors targeting HLA-restricted neoantigens.
Nat Commun, 12, 2021
|
|
7JUL
 
 | | Crystal structure of non phosphorylated PTEN (n-crPTEN-13sp-T1, SDTTDSDPENEG) | | Descriptor: | Phosphatidylinositol 3,4,5-trisphosphate 3-phosphatase and dual-specificity protein phosphatase PTEN | | Authors: | Dempsey, D, Phan, K, Cole, P, Gabelli, S.B. | | Deposit date: | 2020-08-20 | | Release date: | 2021-08-11 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.53 Å) | | Cite: | The structural basis of PTEN regulation by multi-site phosphorylation.
Nat.Struct.Mol.Biol., 28, 2021
|
|
2RD0
 
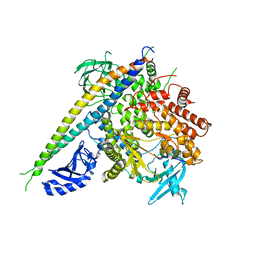 | | Structure of a human p110alpha/p85alpha complex | | Descriptor: | Phosphatidylinositol 3-kinase regulatory subunit alpha, Phosphatidylinositol-4,5-bisphosphate 3-kinase catalytic subunit alpha isoform | | Authors: | Huang, C, Gabelli, S.B, Amzel, L.M. | | Deposit date: | 2007-09-20 | | Release date: | 2007-12-25 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (3.05 Å) | | Cite: | The structure of a human p110alpha/p85alpha complex elucidates the effects of oncogenic PI3Kalpha mutations.
Science, 318, 2007
|
|
8E0W
 
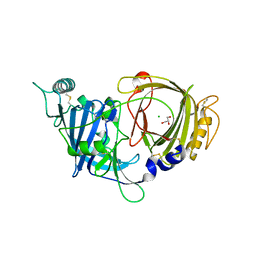 | | Crystal structure of mouse APCDD1 in P1 space group | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CHLORIDE ION, GLYCEROL, ... | | Authors: | Hsieh, F.L, Chang, T.H, Gabelli, S.B, Nathans, J. | | Deposit date: | 2022-08-09 | | Release date: | 2023-05-10 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Structure of WNT inhibitor adenomatosis polyposis coli down-regulated 1 (APCDD1), a cell-surface lipid-binding protein.
Proc.Natl.Acad.Sci.USA, 120, 2023
|
|
8E0R
 
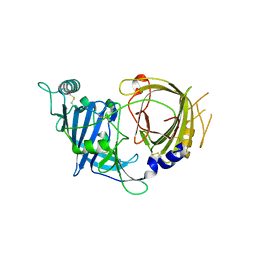 | | Crystal structure of mouse APCDD1 in P21 space group | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Protein APCDD1 | | Authors: | Hsieh, F.L, Chang, T.H, Gabelli, S.B, Nathans, J. | | Deposit date: | 2022-08-09 | | Release date: | 2023-05-10 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Structure of WNT inhibitor adenomatosis polyposis coli down-regulated 1 (APCDD1), a cell-surface lipid-binding protein.
Proc.Natl.Acad.Sci.USA, 120, 2023
|
|
6VJC
 
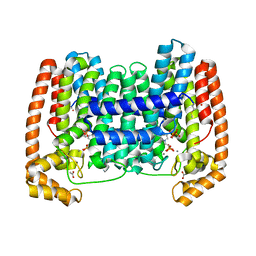 | | LmFPPS mutant T164Y in complex with 476A, IPP & Ca | | Descriptor: | 3-butyl-1-(2,2-diphosphonoethyl)pyridinium, ACETATE ION, CALCIUM ION, ... | | Authors: | Maheshwari, S, Kim, Y.S, Aripirala, S, Gabelli, S.B. | | Deposit date: | 2020-01-15 | | Release date: | 2020-11-25 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Identifying Structural Determinants of Product Specificity in Leishmania major Farnesyl Diphosphate Synthase.
Biochemistry, 59, 2020
|
|
2BRA
 
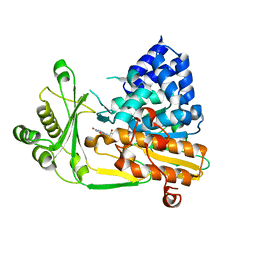 | | Structure of N-Terminal FAD Binding motif of mouse MICAL | | Descriptor: | CHLORIDE ION, FLAVIN-ADENINE DINUCLEOTIDE, NEDD9 INTERACTING PROTEIN WITH CALPONIN HOMOLOGY AND LIM DOMAINS | | Authors: | Nadella, M, Bianchet, M.A, Gabelli, S.B, Amzel, L.M. | | Deposit date: | 2005-05-04 | | Release date: | 2005-11-01 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure and activity of the axon guidance protein MICAL.
Proc. Natl. Acad. Sci. U.S.A., 102, 2005
|
|
6B09
 
 | |
6UUF
 
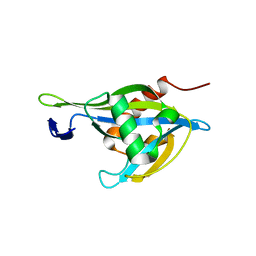 | | Crystal structure of a Nudix Hydrolase from M. Smegmatis, RenU | | Descriptor: | Nudix Hydrolase, RenU | | Authors: | Wright, K.M, Yoder, J, Shoemaker, S, Hernandez, A, Iheanacho, A, Marques, I, Amzel, M.L, Gabelli, S.B. | | Deposit date: | 2019-10-30 | | Release date: | 2021-05-12 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structure of RenU
To Be Published
|
|
3IBA
 
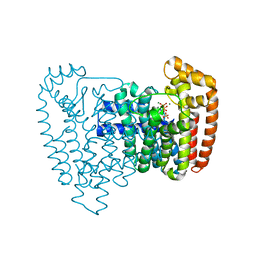 | | Crystal structure of the complex of Trypanosoma cruzi farnesyl diphosphate synthase with zoledronate, IPP and Mg2+ | | Descriptor: | 3-METHYLBUT-3-ENYL TRIHYDROGEN DIPHOSPHATE, Farnesyl pyrophosphate synthase, MAGNESIUM ION, ... | | Authors: | Amzel, L.M, Huang, C.H, Gabelli, S.B, Oldfield, E. | | Deposit date: | 2009-07-15 | | Release date: | 2010-02-09 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Binding of nitrogen-containing bisphosphonates (N-BPs) to the Trypanosoma cruzi farnesyl diphosphate synthase homodimer.
Proteins, 78, 2010
|
|
5W6X
 
 | |
