3ZW0
 
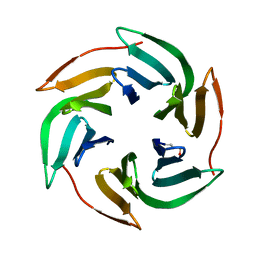 | | Structure of BambL lectin from Burkholderia ambifaria | | Descriptor: | BAMBL LECTIN, alpha-L-fucopyranose | | Authors: | Audfray, A, Claudinon, J, Abounit, S, Ruvoen-Clouet, N, Larson, G, Wimmerova, M, LePendu, J, Romer, W, Varrot, A, Imberty, A. | | Deposit date: | 2011-07-28 | | Release date: | 2011-12-21 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Fucose-Binding Lectin from Opportunistic Pathogen Burkholderia Ambifaria Binds to Both Plant and Human Oligosaccharidic Epitopes.
J.Biol.Chem., 287, 2012
|
|
3ZZV
 
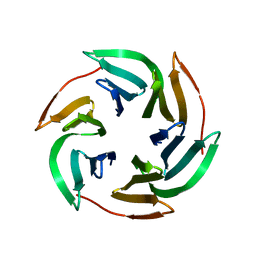 | | BambL complexed with Htype2 tetrasaccharide | | Descriptor: | BAMBL LECTIN, alpha-L-fucopyranose, alpha-L-fucopyranose-(1-2)-beta-D-galactopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose | | Authors: | Audfray, A, Claudinon, J, Abounit, S, Ruvoen-Clouet, N, Larson, G, Wimmerova, M, LePendu, J, Romer, W, Varrot, A, Imberty, A. | | Deposit date: | 2011-09-05 | | Release date: | 2011-12-21 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.68 Å) | | Cite: | Fucose-Binding Lectin from Opportunistic Pathogen Burkholderia Ambifaria Binds to Both Plant and Human Oligosaccharidic Epitopes.
J.Biol.Chem., 287, 2012
|
|
6PVJ
 
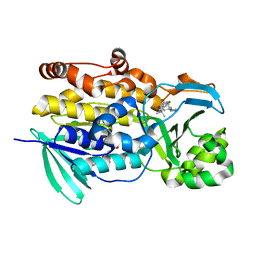 | | Crystal structure of PhqK in complex with malbrancheamide C | | Descriptor: | (5aS,12aS,13aS)-9-bromo-12,12-dimethyl-2,3,11,12,12a,13-hexahydro-1H,5H,6H-5a,13a-(epiminomethano)indolizino[7,6-b]carbazol-14-one, FAD monooxygenase, FLAVIN-ADENINE DINUCLEOTIDE | | Authors: | Fraley, A.E, Smith, J.L, Sherman, D.H. | | Deposit date: | 2019-07-20 | | Release date: | 2020-01-22 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.25 Å) | | Cite: | Molecular Basis for Spirocycle Formation in the Paraherquamide Biosynthetic Pathway.
J.Am.Chem.Soc., 142, 2020
|
|
4ALB
 
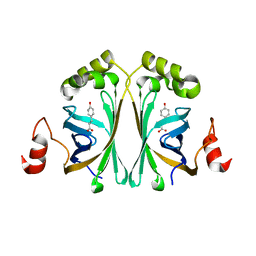 | | Structure of Phenolic Acid Decarboxylase from Bacillus subtilis: Tyr19Ala mutant in complex with coumaric acid | | Descriptor: | 4'-HYDROXYCINNAMIC ACID, PHENOLIC ACID DECARBOXYLASE PADC | | Authors: | Frank, A, Eborall, W, Hyde, R, Hart, S, Turkenburg, J.P, Grogan, G. | | Deposit date: | 2012-03-02 | | Release date: | 2012-08-22 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (3.03 Å) | | Cite: | Mutational Analysis of Phenolic Acid Decarboxylase from Bacillus Subtilis (Bspad), which Converts Bio-Derived Phenolic Acids to Styrene Derivatives
Catal.Sci.Technol., 2, 2012
|
|
6I1O
 
 | | Fab fragment of an antibody selective for wild-type alpha-1-antitrypsin | | Descriptor: | 2-[BIS-(2-HYDROXY-ETHYL)-AMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, FAB 2H2 heavy chain, FAB 2H2 light chain, ... | | Authors: | Laffranchi, M, Elliston, E.L.K, Miranda, E, Perez, J, Fra, A, Lomas, D.A, Irving, J.A. | | Deposit date: | 2018-10-29 | | Release date: | 2019-11-20 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.93 Å) | | Cite: | Intrahepatic heteropolymerization of M and Z alpha-1-antitrypsin.
JCI Insight, 5, 2020
|
|
6I3Z
 
 | | Fab fragment of an antibody selective for wild-type alpha-1-antitrypsin in complex with its antigen | | Descriptor: | Alpha-1-antitrypsin, Fab 2H2 heavy chain, Fab 2H2 light chain, ... | | Authors: | Laffranchi, M, Elliston, E.L.K, Miranda, E, Perez, J, Jagger, A.M, Fra, A, Lomas, D.A, Irving, J.A. | | Deposit date: | 2018-11-08 | | Release date: | 2019-11-20 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Intrahepatic heteropolymerization of M and Z alpha-1-antitrypsin.
JCI Insight, 5, 2020
|
|
6W1X
 
 | | Cryo-EM structure of anti-CRISPR AcrIF9, bound to the type I-F crRNA-guided CRISPR surveillance complex | | Descriptor: | CRISPR-associated endonuclease Cas6/Csy4, CRISPR-associated protein Csy1, CRISPR-associated protein Csy3, ... | | Authors: | Hirschi, M, Santiago-Frangos, A, Wilkinson, R, Golden, S.M, Wiedenheft, B, Lander, G. | | Deposit date: | 2020-03-04 | | Release date: | 2020-05-13 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | AcrIF9 tethers non-sequence specific dsDNA to the CRISPR RNA-guided surveillance complex.
Nat Commun, 11, 2020
|
|
4IJH
 
 | | Fragment-based Discovery of Protein-Protein Interaction Inhibitors of Replication Protein A | | Descriptor: | 3-chloro-6-[3-(4-fluorophenyl)-5-sulfanyl-4H-1,2,4-triazol-4-yl]-1-benzothiophene-2-carboxylic acid, Replication protein A 70 kDa DNA-binding subunit | | Authors: | Feldkamp, M.D, Patrone, J.D, Kennedy, J.P, Frank, A.O, Vangamudi, B, Pelz, N.F, Rossanese, O.W, Waterson, A.G, Fesik, S.W, Chazin, W.J. | | Deposit date: | 2012-12-21 | | Release date: | 2013-08-14 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.498 Å) | | Cite: | Discovery of Protein-Protein Interaction Inhibitors of Replication Protein A.
ACS MED.CHEM.LETT., 4, 2013
|
|
4IJL
 
 | | Fragment-based Discovery of Protein-Protein Interaction Inhibitors of Replication Protein A | | Descriptor: | Replication protein A 70 kDa DNA-binding subunit, {[5-(3-chloro-1-benzothiophen-2-yl)-4-phenyl-4H-1,2,4-triazol-3-yl]sulfanyl}acetic acid | | Authors: | Feldkamp, M.D, Patrone, J.D, Kennedy, J.P, Frank, A.O, Vangamudi, B, Pelz, N.F, Rossanese, O.W, Waterson, A.G, Fesik, S.W, Chazin, W.J. | | Deposit date: | 2012-12-21 | | Release date: | 2013-08-14 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Discovery of Protein-Protein Interaction Inhibitors of Replication Protein A.
ACS MED.CHEM.LETT., 4, 2013
|
|
3ZI8
 
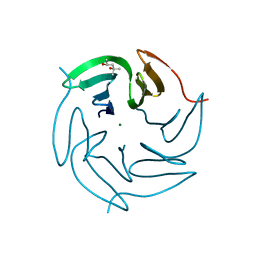 | | Structure of the R17A mutant of the Ralstonia soleanacerum lectin at 1.5 Angstrom in complex with L-fucose | | Descriptor: | (4R)-2-METHYLPENTANE-2,4-DIOL, MAGNESIUM ION, PUTATIVE FUCOSE-BINDING LECTIN PROTEIN, ... | | Authors: | Arnaud, J, Audfray, A, Claudinon, J, Trondle, K, Trosvalet, M, Thomas, A, Varrot, A, Romer, W, Imberty, A. | | Deposit date: | 2013-01-07 | | Release date: | 2013-07-31 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Reduction of Lectin Valency Drastically Changes Glycolipid Dynamics in Membranes, But not Surface Avidity.
Acs Chem.Biol., 8, 2013
|
|
4IPC
 
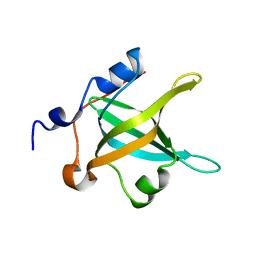 | | Structure of the N-terminal domain of RPA70, E7R mutant | | Descriptor: | Replication protein A 70 kDa DNA-binding subunit | | Authors: | Feldkamp, M.D, Frank, A.O, Vangamudi, B, Fesik, S.W, Chazin, W.J. | | Deposit date: | 2013-01-09 | | Release date: | 2013-09-11 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.22 Å) | | Cite: | Surface Reengineering of RPA70N Enables Cocrystallization with an Inhibitor of the Replication Protein A Interaction Motif of ATR Interacting Protein.
Biochemistry, 52, 2013
|
|
4IPD
 
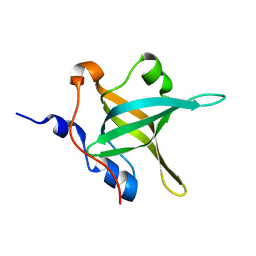 | | Structure of the N-terminal domain of RPA70, E100R mutant | | Descriptor: | Replication protein A 70 kDa DNA-binding subunit | | Authors: | Feldkamp, M.D, Frank, A.O, Vangamudi, B, Fesik, S.W, Chazin, W.J. | | Deposit date: | 2013-01-09 | | Release date: | 2013-09-11 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.51 Å) | | Cite: | Surface Reengineering of RPA70N Enables Cocrystallization with an Inhibitor of the Replication Protein A Interaction Motif of ATR Interacting Protein.
Biochemistry, 52, 2013
|
|
4IPG
 
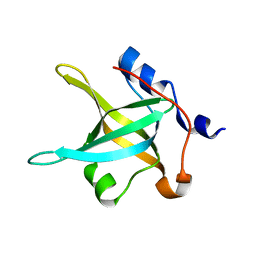 | | Structure of the N-terminal domain of RPA70, E7R, E100R mutant | | Descriptor: | Replication protein A 70 kDa DNA-binding subunit | | Authors: | Feldkamp, M.D, Frank, A.O, Vangamudi, B, Fesik, S.W, Chazin, W.J. | | Deposit date: | 2013-01-09 | | Release date: | 2013-09-11 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.58 Å) | | Cite: | Surface Reengineering of RPA70N Enables Cocrystallization with an Inhibitor of the Replication Protein A Interaction Motif of ATR Interacting Protein.
Biochemistry, 52, 2013
|
|
4IPH
 
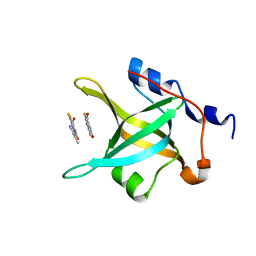 | | Structure of N-terminal domain of RPA70 in complex with VU079104 inhibitor | | Descriptor: | Replication protein A 70 kDa DNA-binding subunit, ~{N}-(2,3-dimethylphenyl)-7-oxidanylidene-12-sulfanylidene-5,11-dithia-1,8-diazatricyclo[7.3.0.0^{2,6}]dodeca-2(6),3,9-triene-10-carboxamide | | Authors: | Feldkamp, M.D, Frank, A.O, Vangamudi, B, Fesik, S.W, Chazin, W.J. | | Deposit date: | 2013-01-09 | | Release date: | 2013-09-11 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.94 Å) | | Cite: | Surface Reengineering of RPA70N Enables Cocrystallization with an Inhibitor of the Replication Protein A Interaction Motif of ATR Interacting Protein.
Biochemistry, 52, 2013
|
|
3ZGY
 
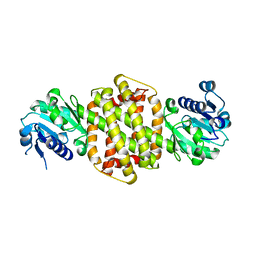 | |
4R4I
 
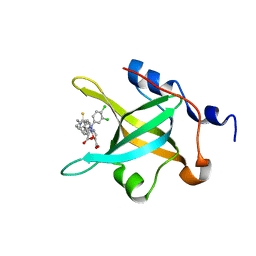 | | Structure of RPA70N in complex with 5-(4-((6-(5-carboxyfuran-2-yl)-1-thioxo-3,4-dihydroisoquinolin-2(1H)-yl)methyl)phenyl)-1-(3,4-dichlorophenyl)-1H-pyrazole-3-carboxylic acid | | Descriptor: | 5-(4-{[6-(5-carboxyfuran-2-yl)-1-thioxo-3,4-dihydroisoquinolin-2(1H)-yl]methyl}phenyl)-1-(3,4-dichlorophenyl)-1H-pyrazole-3-carboxylic acid, Replication protein A 70 kDa DNA-binding subunit | | Authors: | Feldkamp, M.D, Waterson, A.G, Kennedy, J.P, Patrone, J.D, Pelz, N.F, Frank, A.O, Vangamudi, B, Sousa-Fagundes, E.M, Rossanese, O.W, Fesik, S.W, Chazin, W.J. | | Deposit date: | 2014-08-19 | | Release date: | 2014-11-19 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Diphenylpyrazoles as replication protein a inhibitors.
ACS Med Chem Lett, 6, 2015
|
|
4R4O
 
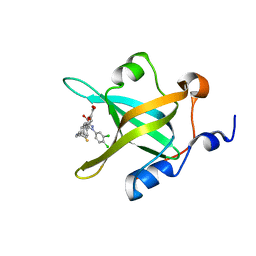 | | Crystal structure of RPA70N in complex with 5-(4-((4-(5-carboxyfuran-2-yl)benzyl)carbamothioyl)phenyl)-1-(3,4-dichlorophenyl)-1H-pyrazole-3-carboxylic acid | | Descriptor: | 5-(4-{[4-(5-carboxyfuran-2-yl)benzyl]carbamothioyl}phenyl)-1-(3,4-dichlorophenyl)-1H-pyrazole-3-carboxylic acid, Replication protein A 70 kDa DNA-binding subunit | | Authors: | Feldkamp, M.D, Waterson, A.G, Kennedy, J.P, Patrone, J.D, Pelz, N.F, Frank, A.O, Vangamudi, B, Sousa-Fagundes, E.M, Rossanese, O.W, Fesik, S.W, Chazin, W.J. | | Deposit date: | 2014-08-19 | | Release date: | 2014-11-19 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.33 Å) | | Cite: | Diphenylpyrazoles as replication protein a inhibitors.
ACS Med Chem Lett, 6, 2015
|
|
4R4T
 
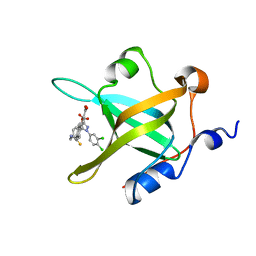 | | Crystal Structure of RPA70N in complex with 5-(4-((4-(5-carboxyfuran-2-yl)phenylthioamido)methyl)phenyl)-1-(3,4-dichlorophenyl)-1H-pyrazole-3-carboxylic acid | | Descriptor: | 5-{4-[({[4-(5-carboxyfuran-2-yl)phenyl]carbonothioyl}amino)methyl]phenyl}-1-(3,4-dichlorophenyl)-1H-pyrazole-3-carboxylic acid, Replication protein A 70 kDa DNA-binding subunit | | Authors: | Feldkamp, M.D, Waterson, A.G, Kennedy, J.P, Patrone, J.D, Pelz, N.F, Frank, A.O, Vangamudi, B, Sousa-Fagundes, E.M, Rossanese, O.W, Fesik, S.W, Chazin, W.J. | | Deposit date: | 2014-08-19 | | Release date: | 2014-11-19 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.28 Å) | | Cite: | Diphenylpyrazoles as replication protein a inhibitors.
ACS Med Chem Lett, 6, 2015
|
|
4R4Q
 
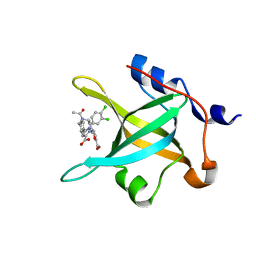 | | Crystal structure of RPA70N in complex with 5-(3-((N-(4-(5-carboxyfuran-2-yl)benzyl)acetamido)methyl)phenyl)-1-(3,4-dichlorophenyl)-1H-pyrazole-3-carboxylic acid | | Descriptor: | 5-[4-({acetyl[4-(5-carboxyfuran-2-yl)benzyl]amino}methyl)phenyl]-1-(3,4-dichlorophenyl)-1H-pyrazole-3-carboxylic acid, Replication protein A 70 kDa DNA-binding subunit | | Authors: | Feldkamp, M.D, Waterson, A.G, Kennedy, J.P, Patrone, J.D, Pelz, N.F, Frank, A.O, Vangamudi, B, Sousa-Fagundes, E.M, Rossanese, O.W, Fesik, S.W, Chazin, W.J. | | Deposit date: | 2014-08-19 | | Release date: | 2014-11-19 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Diphenylpyrazoles as replication protein a inhibitors.
ACS Med Chem Lett, 6, 2015
|
|
4R4C
 
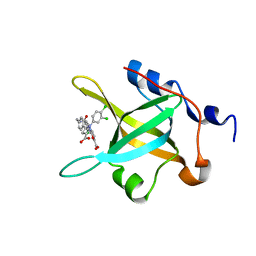 | | Structure of RPA70N in complex with 5-(4-((4-(5-carboxyfuran-2-yl)-2-chlorobenzamido)methyl)phenyl)-1-(3,4-dichlorophenyl)-1H-pyrazole-3-carboxylic acid | | Descriptor: | 5-[4-({[4-(5-carboxyfuran-2-yl)-2-chlorobenzoyl]amino}methyl)phenyl]-1-(3,4-dichlorophenyl)-1H-pyrazole-3-carboxylic acid, Replication protein A 70 kDa DNA-binding subunit | | Authors: | Feldkamp, M.D, Waterson, A.G, Kennedy, J.P, Patrone, J.D, Pelz, N.F, Frank, A.O, Vangamudi, B, Sousa-Fagundes, E.M, Rossanese, O.W, Fesik, S.W, Chazin, W.J. | | Deposit date: | 2014-08-19 | | Release date: | 2014-11-19 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Diphenylpyrazoles as replication protein a inhibitors.
ACS Med Chem Lett, 6, 2015
|
|
4S39
 
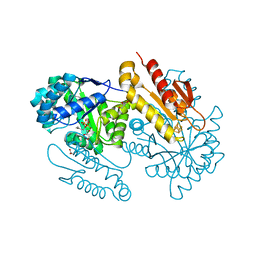 | | IspG in complex with HMBPP | | Descriptor: | (2E)-4-hydroxy-3-methylbut-2-en-1-yl trihydrogen diphosphate, 4-hydroxy-3-methylbut-2-en-1-yl diphosphate synthase, GLYCEROL, ... | | Authors: | Quitterer, F, Frank, A, Wang, K, Guodong, R, O'Dowd, B, Li, J, Guerra, F, Abdel-Azeim, S, Bacher, A, Eppinger, J, Oldfield, E, Groll, M. | | Deposit date: | 2015-01-26 | | Release date: | 2015-04-29 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Atomic-Resolution Structures of Discrete Stages on the Reaction Coordinate of the [Fe4S4] Enzyme IspG (GcpE).
J.Mol.Biol., 427, 2015
|
|
6WIV
 
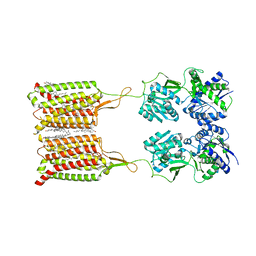 | | Structure of human GABA(B) receptor in an inactive state | | Descriptor: | (2R)-3-{[(S)-(2-aminoethoxy)(hydroxy)phosphoryl]oxy}-2-{[(9Z)-octadec-9-enoyl]oxy}propyl (5Z,8Z,11Z,14Z)-icosa-5,8,11,14-tetraenoate, 2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, ... | | Authors: | Park, J, Fu, Z, Frangaj, A, Liu, J, Mosyak, L, Shen, T, Slavkovich, V.N, Ray, K.M, Taura, J, Cao, B, Geng, Y, Zuo, H, Kou, Y, Grassucci, R, Chen, S, Liu, Z, Lin, X, Williams, J.P, Rice, W.J, Eng, E.T, Huang, R.K, Soni, R.K, Kloss, B, Yu, Z, Javitch, J.A, Hendrickson, W.A, Slesinger, P.A, Quick, M, Graziano, J, Yu, H, Fiehn, O, Clarke, O.B, Frank, J, Fan, Q.R. | | Deposit date: | 2020-04-10 | | Release date: | 2020-07-01 | | Last modified: | 2020-08-26 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Structure of human GABABreceptor in an inactive state.
Nature, 584, 2020
|
|
9AVL
 
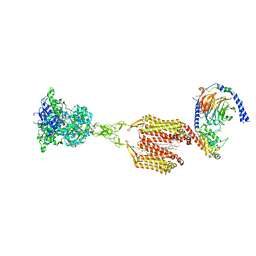 | | Structure of human calcium-sensing receptor in complex with Gi3 protein in nanodiscs | | Descriptor: | (19R,22S,25R)-22,25,26-trihydroxy-16,22-dioxo-17,21,23-trioxa-22lambda~5~-phosphahexacosan-19-yl (9Z)-octadec-9-enoate, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Zuo, H, Park, J, Frangaj, A, Ye, J, Lu, G, Manning, J.J, Asher, W.B, Lu, Z, Hu, G, Wang, L, Mendez, J, Eng, E, Zhang, Z, Lin, X, Grasucci, R, Hendrickson, W.A, Clarke, O.B, Javitch, J.A, Conigrave, A.D, Fan, Q.R. | | Deposit date: | 2024-03-04 | | Release date: | 2024-04-17 | | Last modified: | 2024-05-22 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Promiscuous G-protein activation by the calcium-sensing receptor.
Nature, 629, 2024
|
|
9AYF
 
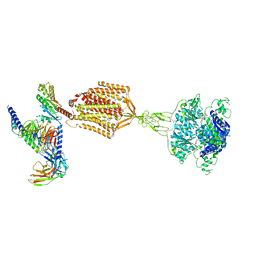 | | Structure of human calcium-sensing receptor in complex with Gi1 (miniGi1) protein in detergent | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 3-(2-chlorophenyl)-N-[(1R)-1-(3-methoxyphenyl)ethyl]propan-1-amine, ... | | Authors: | Zuo, H, Park, J, Frangaj, A, Ye, J, Lu, G, Manning, J.J, Asher, W.B, Lu, Z, Hu, G, Wang, L, Mendez, J, Eng, E, Zhang, Z, Lin, X, Grasucci, R, Hendrickson, W.A, Clarke, O.B, Javitch, J.A, Conigrave, A.D, Fan, Q.R. | | Deposit date: | 2024-03-07 | | Release date: | 2024-04-17 | | Last modified: | 2024-05-22 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Promiscuous G-protein activation by the calcium-sensing receptor.
Nature, 629, 2024
|
|
9AVG
 
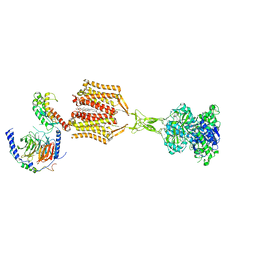 | | Structure of human calcium-sensing receptor in complex with chimeric Gs (miniGis) protein in nanodiscs | | Descriptor: | (19R,22S,25R)-22,25,26-trihydroxy-16,22-dioxo-17,21,23-trioxa-22lambda~5~-phosphahexacosan-19-yl (9Z)-octadec-9-enoate, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Zuo, H, Park, J, Frangaj, A, Ye, J, Lu, G, Manning, J.J, Asher, W.B, Lu, Z, Hu, G, Wang, L, Mendez, J, Eng, E, Zhang, Z, Lin, X, Grasucci, R, Hendrickson, W.A, Clarke, O.B, Javitch, J.A, Conigrave, A.D, Fan, Q.R. | | Deposit date: | 2024-03-02 | | Release date: | 2024-04-17 | | Last modified: | 2024-05-22 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Promiscuous G-protein activation by the calcium-sensing receptor.
Nature, 629, 2024
|
|
