2PSB
 
 | | Crystal structure of YerB protein from Bacillus subtilis. NorthEast Structural Genomics target SR586 | | Descriptor: | YerB protein | | Authors: | Seetharaman, J, Chen, Y, Forouhar, F, Wang, D, Fang, Y, Cunningham, K, Ma, L.-C, Xia, R, Liu, J, Baran, M.C, Acton, T.B, Rost, B, Montelione, G.T, Hunt, J.F, Tong, L, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2007-05-04 | | Release date: | 2007-05-15 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structure of YerB protein from Bacillus subtilis.
To be Published
|
|
3THT
 
 | | Crystal structure and RNA binding properties of the RRM/AlkB domains in human ABH8, an enzyme catalyzing tRNA hypermodification, Northeast Structural Genomics Consortium Target HR5601B | | Descriptor: | 2-OXOGLUTARIC ACID, Alkylated DNA repair protein alkB homolog 8, MANGANESE (II) ION, ... | | Authors: | Pastore, C, Topalidou, I, Forouhar, F, Yan, A.C, Levy, M, Hunt, J.F, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2011-08-19 | | Release date: | 2011-11-02 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (3.01 Å) | | Cite: | Crystal structure and RNA binding properties of the RNA recognition motif (RRM) and AlkB domains in human AlkB homolog 8 (ABH8), an enzyme catalyzing tRNA hypermodification.
J.Biol.Chem., 287, 2012
|
|
3THP
 
 | | Crystal structure and RNA binding properties of the RRM/AlkB domains in human ABH8, an enzyme catalyzing tRNA hypermodification, Northeast Structural Genomics Consortium Target HR5601B | | Descriptor: | 2-OXOGLUTARIC ACID, Alkylated DNA repair protein alkB homolog 8, MANGANESE (II) ION, ... | | Authors: | Pastore, C, Topalidou, I, Forouhar, F, Yan, A.C, Levy, M, Hunt, J.F, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2011-08-19 | | Release date: | 2011-11-02 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Crystal structure and RNA binding properties of the RNA recognition motif (RRM) and AlkB domains in human AlkB homolog 8 (ABH8), an enzyme catalyzing tRNA hypermodification.
J.Biol.Chem., 287, 2012
|
|
7VYQ
 
 | | Short chain dehydrogenase (SCR) cryoEM structure with NADP and ethyl 4-chloroacetoacetate | | Descriptor: | Carbonyl Reductase, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, ethyl 4-chloranyl-3-oxidanylidene-butanoate | | Authors: | Li, Y.H, Zhang, R.Z, Wang, C, Forouhar, F, Clarke, O, Vorobiev, S, Singh, S, Montelione, G.T, Szyperski, T, Xu, Y, Hunt, J.F. | | Deposit date: | 2021-11-15 | | Release date: | 2022-04-06 | | Last modified: | 2024-06-26 | | Method: | ELECTRON MICROSCOPY (3.13 Å) | | Cite: | Oligomeric interactions maintain active-site structure in a noncooperative enzyme family.
Embo J., 41, 2022
|
|
3BU2
 
 | | Crystal structure of a tRNA-binding protein from Staphylococcus saprophyticus subsp. saprophyticus. Northeast Structural Genomics Consortium target SyR77 | | Descriptor: | Putative tRNA-binding protein | | Authors: | Seetharaman, J, Su, M, Forouhar, F, Wang, D, Fang, Y, Cunningham, K, Ma, L.-C, Xia, R, Liu, J, Baran, M.C, Acton, T.B, Rost, B, Montelione, G.T, Hunt, J.F, Tong, L, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2007-12-31 | | Release date: | 2008-01-22 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Crystal structure of a tRNA-binding protein from Staphylococcus saprophyticus subsp. saprophyticus.
To be Published
|
|
3C37
 
 | | X-ray structure of the putative Zn-dependent peptidase Q74D82 at the resolution 1.7 A. Northeast Structural Genomics Consortium target GsR143A | | Descriptor: | DI(HYDROXYETHYL)ETHER, Peptidase, M48 family, ... | | Authors: | Kuzin, A.P, Chen, Y, Seetharaman, J, Vorobiev, S.M, Forouhar, F, Wang, D, Mao, L, Maglaqui, M, Xiao, R, Liu, J, Baran, M.C, Acton, T.B, Rost, B, Montelione, G.T, Tong, L, Hunt, J.F, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2008-01-27 | | Release date: | 2008-02-12 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | X-ray structure of the putative Zn-dependent peptidase Q74D82 at the resolution 1.7 A.
To be Published
|
|
6MRO
 
 | | Crystal structure of methyl transferase from Methanosarcina acetivorans at 1.6 Angstroms resolution, Northeast Structural Genomics Consortium (NESG) Target MvR53. | | Descriptor: | CALCIUM ION, CHLORIDE ION, S-ADENOSYL-L-HOMOCYSTEINE, ... | | Authors: | Singh, S, Forouhar, F, Wang, C, Hunt, J.F, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2018-10-15 | | Release date: | 2018-10-24 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Crystal structure of a methyl transferase from Methanosarcina acetivorans at 1.6 Angstroms resolution.
To Be Published
|
|
2ES7
 
 | | Crystal structure of Q8ZP25 from Salmonella typhimurium LT2. NESG TARGET STR70 | | Descriptor: | putative thiol-disulfide isomerase and thioredoxin | | Authors: | Benach, J, Su, M, Forouhar, F, Chen, Y, Ho, C.K, Janjua, H, Cunningham, K, Ma, L.-C, Rong, X, Liu, J, Baran, M, Acton, T.B, Rost, B, Montelione, G.T, Tong, L, Hunt, J.F, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2005-10-25 | | Release date: | 2005-11-01 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal structure of Q8ZP25 from Salmonella typhimurium LT2 NESG target STR70.
To be Published
|
|
2F20
 
 | | X-ray Crystal Structure of Protein BT_1218 from Bacteroides thetaiotaomicron. Northeast Structural Genomics Consortium Target BtR8. | | Descriptor: | conserved hypothetical protein, with conserved domain | | Authors: | Vorobiev, S.M, Abashidze, M, Seetharaman, J, Forouhar, F, Xiao, R, Ma, L.-C, Montelione, G.T, Tong, L, Hunt, J.F, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2005-11-15 | | Release date: | 2005-11-22 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structure of the Q8A8E9_BACTIN hypothetical protein from Bacteroides thetaiotaomicron.
To be Published
|
|
2FFG
 
 | | Novel x-ray structure of the YkuJ protein from Bacillus subtilis. Northeast Structural Genomics target SR360. | | Descriptor: | ykuJ | | Authors: | Kuzin, A.P, Abashidze, M, Forouhar, F, Vorobiev, S.M, Ho, C.K, Janjua, H, Cunningham, K, Conover, K, Ma, L.C, Xiao, R, Acton, T.B, Montelione, G.T, Tong, L, Hunt, J.F, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2005-12-19 | | Release date: | 2005-12-27 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.31 Å) | | Cite: | Novel x-ray structure of the YkuJ protein from Bacillus subtilis. Northeast Structural Genomics target SR360.
To be Published
|
|
2EW0
 
 | | X-ray Crystal Structure of Protein Q6FF54 from Acinetobacter sp. ADP1. Northeast Structural Genomics Consortium Target AsR1. | | Descriptor: | SULFATE ION, hypothetical protein ACIAD0353 | | Authors: | Kuzin, A.P, Abashidze, M, Vorobiev, S.M, Forouhar, F, Acton, T, Xiao, R, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2005-11-01 | | Release date: | 2005-12-06 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Novel x-ray structure of hypothetical protein Q6FF54 at the resolution 1.4 A. Northeast Structural Genomics target AsR1.
TO BE PUBLISHED
|
|
2F9F
 
 | | Crystal Structure of the Putative Mannosyl Transferase (wbaZ-1)from Archaeoglobus fulgidus, Northeast Structural Genomics Target GR29A. | | Descriptor: | first mannosyl transferase (wbaZ-1) | | Authors: | Zhou, W, Forouhar, F, Conover, K, Xiao, R, Acton, T.B, Montelione, G.T, Tong, L, Hunt, J.F, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2005-12-05 | | Release date: | 2006-06-06 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal Structure of the Putative Mannosyl Transferase
(wbaZ-1)from Archaeoglobus fulgidus
To be Published
|
|
3KKJ
 
 | | X-ray structure of P. syringae q888a4 Oxidoreductase at resolution 2.5A, Northeast Structural Genomics Consortium target PsR10 | | Descriptor: | Amine oxidase, flavin-containing, FLAVIN-ADENINE DINUCLEOTIDE | | Authors: | Kuzin, A.P, Chen, Y, Forouhar, F, Vorobiev, S, Acton, T, Ma, L.C, Xiao, R, Montelione, G.T, Hunt, J.F, Tong, T, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2009-11-05 | | Release date: | 2009-11-24 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | X-ray structure of P. syringae q888a4 Oxidoreductase at resolution 2.5A, Northeast Structural Genomics Consortium target PsR10
To be Published
|
|
1H65
 
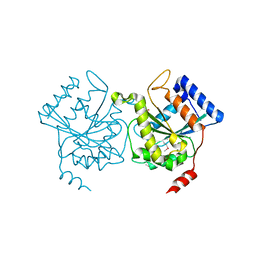 | | Crystal structure of pea Toc34 - a novel GTPase of the chloroplast protein translocon | | Descriptor: | CHLOROPLAST OUTER ENVELOPE PROTEIN OEP34, GUANOSINE-5'-DIPHOSPHATE, MAGNESIUM ION | | Authors: | Sun, Y.J, Forouhar, F, Li, H.M, Tu, S.L, Kao, S, Shr, H.L, Chou, C.C, Hsiao, C.D. | | Deposit date: | 2001-06-06 | | Release date: | 2002-01-29 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal Structure of Pea Toc34 - a Novel Gtpase of the Chloroplast Protein Translocon
Nat.Struct.Biol., 9, 2002
|
|
3KLW
 
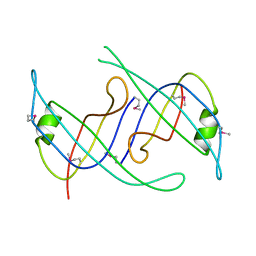 | | Crystal structure of primosomal replication protein n from Bordetella pertussis. northeast structural genomics consortium target ber132. | | Descriptor: | Primosomal replication protein n | | Authors: | Kuzin, A.P, Neely, H, Vorobiev, S.M, Seetharaman, J, Forouhar, F, Wang, H, Janjua, H, Maglaqui, M, Xiao, T, Liu, J, Baran, M.C, Acton, T.B, Rost, B, Montelione, G.T, Hunt, J.F, Tong, L, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2009-11-09 | | Release date: | 2009-11-24 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of primosomal replication protein n from Bordetella pertussis. northeast structural genomics consortium target ber132.
To be Published
|
|
3U1O
 
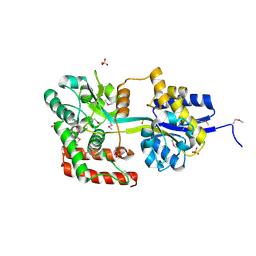 | | THREE DIMENSIONAL STRUCTURE OF DE NOVO DESIGNED CYSTEINE ESTERASE ECH19, Northeast Structural Genomics Consortium Target OR49 | | Descriptor: | De Novo design cysteine esterase ECH19, SODIUM ION, SULFATE ION | | Authors: | Kuzin, A, Su, M, Lew, S, Forouhar, F, Seetharaman, J, Daya, P, Xiao, R, Ciccosanti, C, Richter, F, Everett, J.K, Acton, T.B, Baker, D, Montelione, G.T, Hunt, J.F, Tong, L, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2011-09-30 | | Release date: | 2011-10-26 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (2.494 Å) | | Cite: | Computational design of catalytic dyads and oxyanion holes for ester hydrolysis.
J.Am.Chem.Soc., 134, 2012
|
|
4OHF
 
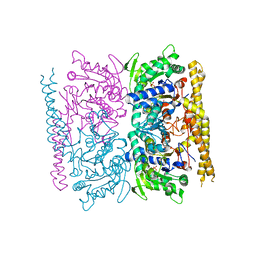 | | Crystal structure of cytosolic nucleotidase II (LPG0095) in complex with GMP from Legionella pneumophila, NORTHEAST STRUCTURAL GENOMICS CONSORTIUM TARGET LGR1 | | Descriptor: | Cytosolic IMP-GMP specific 5'-nucleotidase, GUANOSINE-5'-MONOPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Srinivisan, B, Forouhar, F, Shukla, A, Sampangi, C, Kulkarni, S, Abashidze, M, Seetharaman, J, Lew, S, Mao, L, Acton, T.B, Xiao, R, Everett, J.K, Montelione, G.M, Tong, L, Balaram, H, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2014-01-17 | | Release date: | 2014-02-26 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.53 Å) | | Cite: | Allosteric regulation and substrate activation in cytosolic nucleotidase II from Legionella pneumophila.
Febs J., 281, 2014
|
|
7DN1
 
 | | Hetero-oligomers of SCR-SCR2 crystal structure with NADPH | | Descriptor: | (S)-specific carbonyl reductase, Carbonyl Reductase, NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Li, Y.H, Zhang, R.Z, Forouhar, F, Wang, C, Montelione, G.T, Szyperski, T, Xu, Y, Hunt, J.F. | | Deposit date: | 2020-12-08 | | Release date: | 2022-04-06 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.74 Å) | | Cite: | Oligomeric interactions maintain active-site structure in a noncooperative enzyme family.
Embo J., 41, 2022
|
|
7DLL
 
 | | Short chain dehydrogenase 2 (SCR2) crystal structure with NADPH | | Descriptor: | (S)-specific carbonyl reductase, NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Li, Y.H, Zhang, R.Z, Forouhar, F, Wang, C, Montelione, G.T, Szyperski, T, Xu, Y, Hunt, J.F. | | Deposit date: | 2020-11-28 | | Release date: | 2022-04-06 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.89 Å) | | Cite: | Oligomeric interactions maintain active-site structure in a noncooperative enzyme family.
Embo J., 41, 2022
|
|
7DMG
 
 | | Short chain dehydrogenase 2 (SCR2) crystal structure with NADP | | Descriptor: | (S)-specific carbonyl reductase, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Li, Y.H, Zhang, R.Z, Forouhar, F, Wang, C, Montelione, G.T, Szyperski, T, Xu, Y, Hunt, J.F. | | Deposit date: | 2020-12-03 | | Release date: | 2022-04-06 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.79 Å) | | Cite: | Oligomeric interactions maintain active-site structure in a noncooperative enzyme family.
Embo J., 41, 2022
|
|
7DLM
 
 | | Short chain dehydrogenase (SCR) crystal structure with NADPH | | Descriptor: | Carbonyl Reductase, NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Li, Y.H, Zhang, R.Z, Forouhar, F, Wang, C, Montelione, G.T, Szyperski, T, Xu, Y, Hunt, J.F. | | Deposit date: | 2020-11-28 | | Release date: | 2022-04-06 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.59 Å) | | Cite: | Oligomeric interactions maintain active-site structure in a noncooperative enzyme family.
Embo J., 41, 2022
|
|
7DLD
 
 | | Crystal structures of (S)-carbonyl reductases from Candida parapsilosis in different oligomerization states | | Descriptor: | Carbonyl Reductase, MAGNESIUM ION | | Authors: | Li, Y.H, Zhang, R.Z, Forouhar, F, Wang, C, Montelione, G.T, Szyperski, T, Xu, Y, Hunt, J.F. | | Deposit date: | 2020-11-27 | | Release date: | 2022-04-06 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Oligomeric interactions maintain active-site structure in a noncooperative enzyme family.
Embo J., 41, 2022
|
|
3B57
 
 | | Crystal structure of the Lin1889 protein (Q92AN1) from Listeria innocua. Northeast Structural Consortium target LkR65 | | Descriptor: | Lin1889 protein, MAGNESIUM ION | | Authors: | Kuzin, A.P, Abashidze, M, Seetharaman, J, Benach, J, Forouhar, F, Vorobiev, S.M, Wang, H, Janjua, H, Cunningham, K, Ma, L.-C, Xiao, R, Liu, J, Baran, M.C, Acton, T.B, Rost, B, Montelione, G.T, Tong, L, Hunt, J.F, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2007-10-25 | | Release date: | 2007-11-06 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Crystal structure of the Lin1889 protein (Q92AN1) from Listeria innocua.
To be Published
|
|
3BDR
 
 | | Crystal structure of fatty acid-binding protein-like Ycf58 from Thermosynecoccus elongatus. Northeast Structural Genomics Consortium target TeR13. | | Descriptor: | PHOSPHATE ION, Ycf58 protein | | Authors: | Kuzin, A.P, Su, M, Seetharaman, J, Forouhar, F, Wang, D, Janjua, H, Cunningham, K, Ma, L.-C, Xiao, R, Liu, J, Baran, M.C, Acton, T.B, Rost, B, Montelione, G.T, Tong, L, Hunt, J.F, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2007-11-15 | | Release date: | 2007-11-27 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | The crystal structure of fatty acid-binding protein-like Ycf58 from Thermosynecoccus elongatus.
To be Published
|
|
4ZHN
 
 | | Crystal Structure of AlkB T208A mutant protein in complex with Co(II), 2-oxoglutarate, and methylated trinucleotide T-meA-T | | Descriptor: | 2-OXOGLUTARIC ACID, Alpha-ketoglutarate-dependent dioxygenase AlkB, COBALT (II) ION, ... | | Authors: | Ergel, B, Yu, B, Vorobiev, S.M, Forouhar, F, Hunt, J.F, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2015-04-25 | | Release date: | 2015-12-23 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.333 Å) | | Cite: | DFT Studies of the Rate-Limiting Step in the Reaction Cycle of the Fe/2OG Dioxygenase AlkB and Related Experimental Studies
To be published
|
|
