4LRT
 
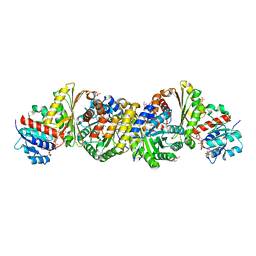 | | Crystal and solution structures of the bifunctional enzyme (Aldolase/Aldehyde dehydrogenase) from Thermomonospora curvata, reveal a cofactor-binding domain motion during NAD+ and CoA accommodation whithin the shared cofactor-binding site | | Descriptor: | 4-hydroxy-2-oxovalerate aldolase, Acetaldehyde dehydrogenase, COENZYME A, ... | | Authors: | Fischer, B, Branlant, G, Talfournier, F, Gruez, A. | | Deposit date: | 2013-07-20 | | Release date: | 2013-09-04 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Crystal and solution structures of the bifunctional enzyme (Aldolase/Aldehyde dehydrogenase) from Thermomonospora curvata, reveal a cofactor-binding domain motion during NAD+ and CoA accommodation whithin the shared cofactor-binding site
To be Published
|
|
5HJQ
 
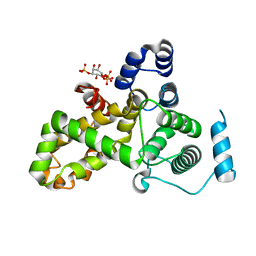 | | Crystal structure of the TBC domain of Skywalker/TBC1D24 from Drosophila melanogaster in complex with inositol(1,4,5)triphosphate | | Descriptor: | D-MYO-INOSITOL-1,4,5-TRIPHOSPHATE, LD10117p | | Authors: | Fischer, B, Paesmans, J, Versees, W. | | Deposit date: | 2016-01-13 | | Release date: | 2016-09-21 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Skywalker-TBC1D24 has a lipid-binding pocket mutated in epilepsy and required for synaptic function.
Nat.Struct.Mol.Biol., 23, 2016
|
|
5HJN
 
 | |
6R82
 
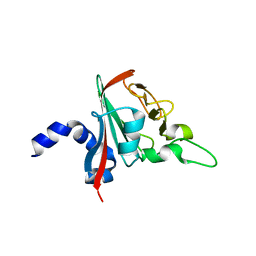 | |
8BE8
 
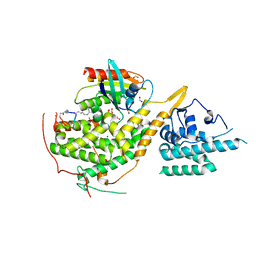 | | Crystal structure of SOS1-HRas-peptidomimetic4 | | Descriptor: | FORMIC ACID, GTPase HRas, SOS1-HRas-peptidomimetic4, ... | | Authors: | Fischer, B, Wohlkonig, A, Steyaert, J. | | Deposit date: | 2022-10-21 | | Release date: | 2023-04-26 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Nanobody Loop Mimetics Enhance Son of Sevenless 1-Catalyzed Nucleotide Exchange on RAS.
Angew.Chem.Int.Ed.Engl., 62, 2023
|
|
8BE9
 
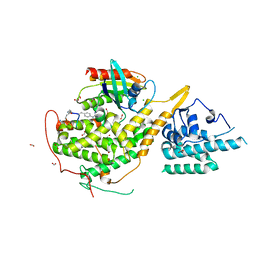 | | Crystal structure of SOS1-HRas-peptidomimetic5 | | Descriptor: | CHLORIDE ION, FORMIC ACID, GTPase HRas, ... | | Authors: | Fischer, B, Wohlkonig, A, Steyaert, J. | | Deposit date: | 2022-10-21 | | Release date: | 2023-04-26 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.51 Å) | | Cite: | Nanobody Loop Mimetics Enhance Son of Sevenless 1-Catalyzed Nucleotide Exchange on RAS.
Angew.Chem.Int.Ed.Engl., 62, 2023
|
|
8BE6
 
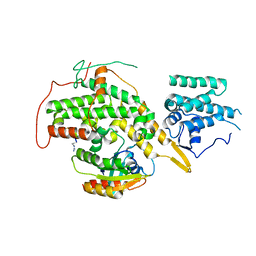 | | Crystal structure of SOS1-HRas-peptidomimetic2 | | Descriptor: | GTPase HRas, SOS1-HRas-peptidomimetic2, Son of sevenless homolog 1 | | Authors: | Fischer, B, Wohlkonig, A, Steyaert, J. | | Deposit date: | 2022-10-21 | | Release date: | 2023-04-26 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.89880252 Å) | | Cite: | Nanobody Loop Mimetics Enhance Son of Sevenless 1-Catalyzed Nucleotide Exchange on RAS.
Angew.Chem.Int.Ed.Engl., 62, 2023
|
|
8BEA
 
 | |
8BE7
 
 | |
8BE4
 
 | |
8BE5
 
 | | Crystal structure of SOS1-KRasG12V-Nanobody22-Nanobody75 | | Descriptor: | Isoform 2B of GTPase KRas, Nanobody22, Nanobody75, ... | | Authors: | Fischer, B, Wohlkonig, A, Steyaert, J. | | Deposit date: | 2022-10-21 | | Release date: | 2023-11-01 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (3.13 Å) | | Cite: | Allosteric nanobodies to study the interactions between SOS1 and RAS.
Nat Commun, 15, 2024
|
|
8BE2
 
 | |
8BE3
 
 | | Crystal structure of KRasG12V-Nanobody84 | | Descriptor: | GUANOSINE-5'-DIPHOSPHATE, Isoform 2B of GTPase KRas, MAGNESIUM ION, ... | | Authors: | Fischer, B, Wohlkonig, A, Steyaert, J. | | Deposit date: | 2022-10-21 | | Release date: | 2023-11-01 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Allosteric nanobodies to study the interactions between SOS1 and RAS.
Nat Commun, 15, 2024
|
|
4LRS
 
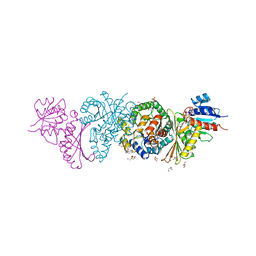 | | Crystal and solution structures of the bifunctional enzyme (Aldolase/Aldehyde dehydrogenase) from Thermomonospora curvata, reveal a cofactor-binding domain motion during NAD+ and CoA accommodation whithin the shared cofactor-binding site | | Descriptor: | 4-hydroxy-2-oxovalerate aldolase, Acetaldehyde dehydrogenase, CHLORIDE ION, ... | | Authors: | Fischer, B, Branlant, G, Talfournier, F, Gruez, A. | | Deposit date: | 2013-07-20 | | Release date: | 2013-09-04 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Crystal and solution structures of the bifunctional enzyme (Aldolase/Aldehyde dehydrogenase) from Thermomonospora curvata, reveal a cofactor-binding domain motion during NAD+ and CoA accommodation whithin the shared cofactor-binding site
To be Published
|
|
6QD6
 
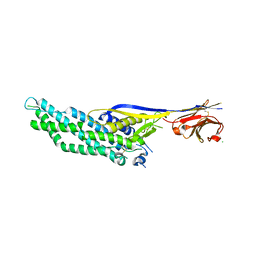 | | Molecular scaffolds expand the nanobody toolkit for cryo-EM applications: crystal structure of Mb-cHopQ-Nb207 | | Descriptor: | CHLORIDE ION, Mb-cHopQ-Nb207,Outer membrane protein,Mb-cHopQ-Nb207,Outer membrane protein,Mb-cHopQ-Nb207 | | Authors: | Uchanski, T, Masiulis, S, Fischer, B, Kalichuk, V, Wohlkonig, A, Zogg, T, Remaut, H, Vranken, W, Aricescu, A.R, Pardon, E, Steyaert, J. | | Deposit date: | 2018-12-31 | | Release date: | 2019-12-18 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.84 Å) | | Cite: | Megabodies expand the nanobody toolkit for protein structure determination by single-particle cryo-EM
Nat.Methods, 18, 2021
|
|
6QFA
 
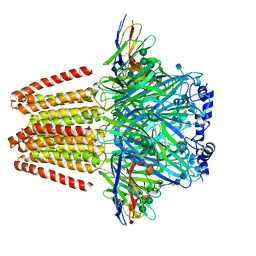 | | CryoEM structure of a beta3K279T GABA(A)R homomer in complex with histamine and megabody Mb25 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Gamma-aminobutyric acid receptor subunit beta-3,Gamma-aminobutyric acid receptor subunit beta-3, HISTAMINE, ... | | Authors: | Uchanski, T, Masiulis, S, Fischer, B, Kalichuk, V, Wohlkoening, A, Zoegg, T, Remaut, H, Vranken, W, Aricescu, A.R, Pardon, E, Steyaert, J. | | Deposit date: | 2019-01-09 | | Release date: | 2021-08-04 | | Last modified: | 2024-10-09 | | Method: | ELECTRON MICROSCOPY (2.49 Å) | | Cite: | Megabodies expand the nanobody toolkit for protein structure determination by single-particle cryo-EM.
Nat.Methods, 18, 2021
|
|
6XUX
 
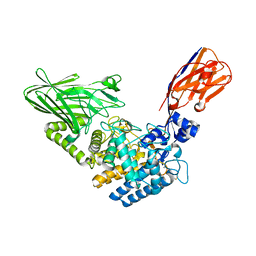 | | Crystal structure of Megabody Mb-Nb207-cYgjK_NO | | Descriptor: | CALCIUM ION, Nanobody,Glucosidase YgjK,Glucosidase YgjK,Nanobody | | Authors: | Steyaert, J, Uchanski, T, Fischer, B. | | Deposit date: | 2020-01-21 | | Release date: | 2021-01-13 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.90000641 Å) | | Cite: | Megabodies expand the nanobody toolkit for protein structure determination by single-particle cryo-EM.
Nat.Methods, 18, 2021
|
|
6XV8
 
 | |
6XVI
 
 | |
