6HLA
 
 | | wild-type NuoEF from Aquifex aeolicus - reduced form bound to NADH | | Descriptor: | 3[N-MORPHOLINO]PROPANE SULFONIC ACID, FE2/S2 (INORGANIC) CLUSTER, FLAVIN MONONUCLEOTIDE, ... | | Authors: | Gerhardt, S, Friedrich, T, Einsle, O, Gnandt, E, Schulte, M, Fiegen, D. | | Deposit date: | 2018-09-11 | | Release date: | 2019-06-26 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | A mechanism to prevent production of reactive oxygen species by Escherichia coli respiratory complex I.
Nat Commun, 10, 2019
|
|
8R4Z
 
 | |
2YIG
 
 | |
8RVC
 
 | | Crystal structure of alpha keto acid C-methyl-transferases MrsA bound to ketoarginine | | Descriptor: | 1,2-ETHANEDIOL, 2-ketoarginine methyltransferase, 5-[(diaminomethylidene)amino]-2-oxopentanoic acid, ... | | Authors: | Gerhardt, S, Kemper, F, Andexer, J.N. | | Deposit date: | 2024-02-01 | | Release date: | 2024-07-03 | | Last modified: | 2024-07-24 | | Method: | X-RAY DIFFRACTION (1.969 Å) | | Cite: | Structures and Protein Engineering of the alpha-Keto Acid C-Methyltransferases SgvM and MrsA for Rational Substrate Transfer.
Chembiochem, 2024
|
|
8RWM
 
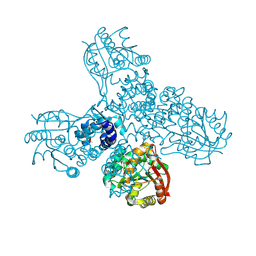 | |
8RWW
 
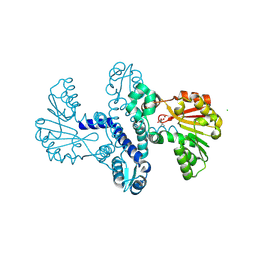 | |
8RVS
 
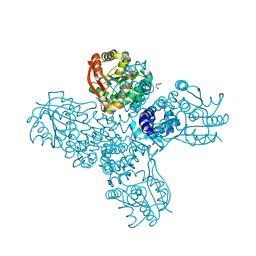 | |
8RXG
 
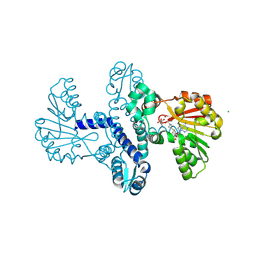 | |
8RXF
 
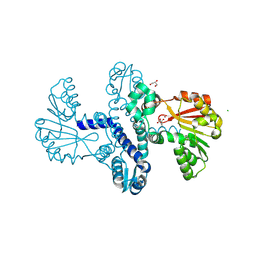 | |
4AA5
 
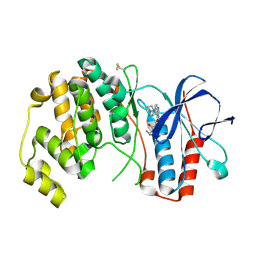 | | P38ALPHA MAP KINASE BOUND TO CMPD 33 | | Descriptor: | MITOGEN-ACTIVATED PROTEIN KINASE 14, N-CYCLOPROPYL-4-METHYL-3-[6-(4-METHYLPIPERAZIN-1-YL)-4-OXIDANYLIDENE-QUINAZOLIN-3-YL]BENZAMIDE | | Authors: | Gerhardt, S, Hargreaves, D. | | Deposit date: | 2011-11-30 | | Release date: | 2012-05-16 | | Last modified: | 2019-02-06 | | Method: | X-RAY DIFFRACTION (2.38 Å) | | Cite: | The Discovery of N-Cyclopropyl-4-Methyl-3-[6--(4-Methylpiperazin-1-Yl-4-Oxoquinazolin-3(4H)-Yl]Benzamide (Azd6703), a Clinical P38Alpha Map Kinase Inhibitor for the Treatment of Inflammatory Diseases
Bioorg.Med.Chem.Lett., 22, 2012
|
|
4AA0
 
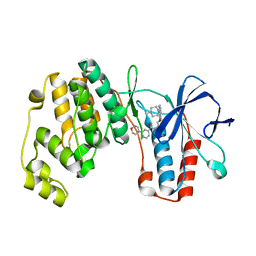 | | P38ALPHA MAP KINASE BOUND TO CMPD 2 | | Descriptor: | MITOGEN-ACTIVATED PROTEIN KINASE 14, N-[4-methyl-3-[6-(4-methylpiperazin-1-yl)-4-oxidanylidene-quinazolin-3-yl]phenyl]-2-morpholin-4-yl-pyridine-4-carboxamide | | Authors: | Gerhardt, S, Hargreaves, D. | | Deposit date: | 2011-11-30 | | Release date: | 2012-05-16 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | The Discovery of N-Cyclopropyl-4-Methyl-3-[6--(4-Methylpiperazin-1-Yl-4-Oxoquinazolin-3(4H)-Yl]Benzamide (Azd6703), a Clinical P38Alpha Map Kinase Inhibitor for the Treatment of Inflammatory Diseases
Bioorg.Med.Chem.Lett., 22, 2012
|
|
4A9Y
 
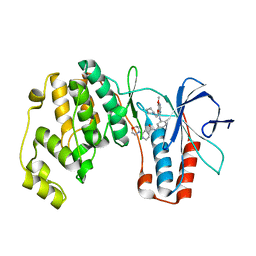 | | P38ALPHA MAP KINASE BOUND TO CMPD 8 | | Descriptor: | MITOGEN-ACTIVATED PROTEIN KINASE 14, N-(3-{[7-METHOXY-6-(2-PYRROLIDIN-1-YLETHOXY)QUINAZOLIN-4-YL]AMINO}-4-METHYLPHENYL)-2-MORPHOLIN-4-YLISONICOTINAMIDE | | Authors: | Gerhardt, S, Hargreaves, D. | | Deposit date: | 2011-11-29 | | Release date: | 2012-05-16 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | The Discovery of N-Cyclopropyl-4-Methyl-3-[6--(4-Methylpiperazin-1-Yl-4-Oxoquinazolin-3(4H)-Yl]Benzamide (Azd6703), a Clinical P38Alpha Map Kinase Inhibitor for the Treatment of Inflammatory Diseases
Bioorg.Med.Chem.Lett., 22, 2012
|
|
4AAC
 
 | | P38ALPHA MAP KINASE BOUND TO CMPD 29 | | Descriptor: | CHLORIDE ION, MITOGEN-ACTIVATED PROTEIN KINASE 14, N-isoxazol-3-yl-4-methyl-3-[6-(4-methylpiperazin-1-yl)-4-oxo-quinazolin-3-yl]benzamide | | Authors: | Gerhardt, S, Hargreaves, D. | | Deposit date: | 2011-12-01 | | Release date: | 2012-05-16 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | The Discovery of N-Cyclopropyl-4-Methyl-3-[6--(4-Methylpiperazin-1-Yl-4-Oxoquinazolin-3(4H)-Yl]Benzamide (Azd6703), a Clinical P38Alpha Map Kinase Inhibitor for the Treatment of Inflammatory Diseases
Bioorg.Med.Chem.Lett., 22, 2012
|
|
4AA4
 
 | | P38ALPHA MAP KINASE BOUND TO CMPD 22 | | Descriptor: | MITOGEN-ACTIVATED PROTEIN KINASE 14, N-[4-METHYL-3-[6-(4-METHYLPIPERAZIN-1-YL)-4-OXIDANYLIDENE-QUINAZOLIN-3-YL]PHENYL]FURAN-3-CARBOXAMIDE | | Authors: | Gerhardt, S, Hargreaves, D. | | Deposit date: | 2011-11-30 | | Release date: | 2012-05-16 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | The Discovery of N-Cyclopropyl-4-Methyl-3-[6--(4-Methylpiperazin-1-Yl-4-Oxoquinazolin-3(4H)-Yl]Benzamide (Azd6703), a Clinical P38Alpha Map Kinase Inhibitor for the Treatment of Inflammatory Diseases
Bioorg.Med.Chem.Lett., 22, 2012
|
|
4JP4
 
 | | Mmp13 in complex with a reverse hydroxamate Zn-binder | | Descriptor: | CALCIUM ION, Collagenase 3, N-[(2S)-4-(5-fluoropyrimidin-2-yl)-1-({4-[5-(2,2,2-trifluoroethoxy)pyrimidin-2-yl]piperazin-1-yl}sulfonyl)butan-2-yl]-N-hydroxyformamide, ... | | Authors: | Gerhardt, S, Hargreaves, D. | | Deposit date: | 2013-03-19 | | Release date: | 2014-03-05 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.43 Å) | | Cite: | Hydantoin based inhibitors of MMP13--discovery of AZD6605.
Bioorg.Med.Chem.Lett., 23, 2013
|
|
4JPA
 
 | | Mmp13 in complex with a piperazine hydantoin ligand | | Descriptor: | 3-[({2-[4-({[(4S)-4-methyl-2,5-dioxoimidazolidin-4-yl]methyl}sulfonyl)piperazin-1-yl]pyrimidin-5-yl}oxy)methyl]benzonitrile, CALCIUM ION, Collagenase 3, ... | | Authors: | Gerhardt, S, Hargreaves, D. | | Deposit date: | 2013-03-19 | | Release date: | 2014-03-05 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Hydantoin based inhibitors of MMP13--discovery of AZD6605.
Bioorg.Med.Chem.Lett., 23, 2013
|
|
2JIH
 
 | | Crystal Structure of Human ADAMTS-1 catalytic Domain and Cysteine- Rich Domain (complex-form) | | Descriptor: | (2S,3R)-N~4~-[(1S)-2,2-dimethyl-1-(methylcarbamoyl)propyl]-N~1~,2-dihydroxy-3-(2-methylpropyl)butanediamide, ADAMTS-1, CADMIUM ION, ... | | Authors: | Gerhardt, S, Hassall, G, Hawtin, P, McCall, E, Flavell, L, Minshull, C, Hargreaves, D, Ting, A, Pauptit, R.A, Parker, A.E, Abbott, W.M. | | Deposit date: | 2007-06-28 | | Release date: | 2008-01-15 | | Last modified: | 2019-04-24 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structures of human ADAMTS-1 reveal a conserved catalytic domain and a disintegrin-like domain with a fold homologous to cysteine-rich domains.
J. Mol. Biol., 373, 2007
|
|
2V4B
 
 | | Crystal Structure of Human ADAMTS-1 catalytic Domain and Cysteine- Rich Domain (apo-form) | | Descriptor: | ADAMTS-1, CADMIUM ION, MAGNESIUM ION, ... | | Authors: | Gerhardt, S, Hassall, G, Hawtin, P, McCall, E, Flavell, L, Minshull, C, Hargreaves, D, Ting, A, Pauptit, R.A, Parker, A.E, Abbott, W.M. | | Deposit date: | 2007-06-28 | | Release date: | 2008-01-15 | | Last modified: | 2019-04-24 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal Structures of Human Adamts-1 Reveal a Conserved Catalytic Domain and a Disintegrin-Like Domain with a Fold Homologous to Cysteine-Rich Domains.
J.Mol.Biol., 373, 2007
|
|
2BAL
 
 | | p38alpha MAP kinase bound to pyrazoloamine | | Descriptor: | Mitogen-activated protein kinase 14, [5-AMINO-1-(4-FLUOROPHENYL)-1H-PYRAZOL-4-YL][3-(PIPERIDIN-4-YLOXY)PHENYL]METHANONE | | Authors: | Gerhardt, S, Pauptit, R.A, Read, J, Tucker, J, Norman, R.A, Breed, J. | | Deposit date: | 2005-10-14 | | Release date: | 2005-12-06 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Prevention of MKK6-Dependent Activation by Binding to p38alpha MAP Kinase.
Biochemistry, 44, 2005
|
|
2BAK
 
 | | p38alpha MAP kinase bound to MPAQ | | Descriptor: | Mitogen-activated protein kinase 14, N-(3-{[7-METHOXY-6-(2-PYRROLIDIN-1-YLETHOXY)QUINAZOLIN-4-YL]AMINO}-4-METHYLPHENYL)-2-MORPHOLIN-4-YLISONICOTINAMIDE | | Authors: | Gerhardt, S, Breed, J, Pauptit, R.A, Read, J, Norman, R.A, Ward, W.H. | | Deposit date: | 2005-10-14 | | Release date: | 2005-12-06 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Prevention of MKK6-Dependent Activation by Binding to p38alpha MAP Kinase
Biochemistry, 44, 2005
|
|
2BAJ
 
 | | p38alpha bound to pyrazolourea | | Descriptor: | 1-(3-tert-butyl-1-phenyl-1H-pyrazol-5-yl)-3-(2,3-dichlorophenyl)urea, Mitogen-activated protein kinase 14 | | Authors: | Gerhardt, S, Pauptit, R.A, Read, J, Breed, J, Norman, R.A, Ward, W.H. | | Deposit date: | 2005-10-14 | | Release date: | 2005-12-06 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Prevention of MKK6-Dependent Activation by Binding to p38alpha MAP Kinase
Biochemistry, 44, 2005
|
|
2BAQ
 
 | | p38alpha bound to Ro3201195 | | Descriptor: | Mitogen-activated protein kinase 14, [5-AMINO-1-(4-FLUOROPHENYL)-1H-PYRAZOL-4-YL](3-{[(2R)-2,3-DIHYDROXYPROPYL]OXY}PHENYL)METHANONE | | Authors: | Gerhardt, S, Pauptit, R.A, Breed, J, Read, J, Tucker, J, Norman, R.A. | | Deposit date: | 2005-10-14 | | Release date: | 2005-12-06 | | Last modified: | 2021-10-20 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Prevention of MKK6-Dependent Activation by Binding to p38alpha MAP Kinase.
Biochemistry, 44, 2005
|
|
2L45
 
 | | C-terminal zinc knuckle of the HIVNCp7 with DNA | | Descriptor: | C-TERMINAL ZINC KNUCLE OF THE HIV-NCP7, DNA (5'-D(P*TP*AP*CP*GP*CP*C)-3'), ZINC ION | | Authors: | Quintal, S, Viegas, A, Cabrita, E, Farrell, N, Erhardt, S. | | Deposit date: | 2010-10-01 | | Release date: | 2011-12-14 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Platinated DNA affects zinc finger conformation. Interaction of a platinated single-stranded oligonucleotide and the C-terminal zinc finger of nucleocapsid protein HIVNCp7.
Biochemistry, 51, 2012
|
|
2L46
 
 | | C-terminal zinc finger of the HIVNCp7 with platinated DNA | | Descriptor: | C-TERMINAL ZINC KNUCLE OF THE HIV-NCP7, DNA (5'-D(P*TP*AP*CP*GP*CP*C)-3'), ZINC ION | | Authors: | Quintal, S, Viegas, A, Cabrita, E, Farrell, N, Erhardt, S. | | Deposit date: | 2010-10-01 | | Release date: | 2011-12-14 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Platinated DNA affects zinc finger conformation. Interaction of a platinated single-stranded oligonucleotide and the C-terminal zinc finger of nucleocapsid protein HIVNCp7.
Biochemistry, 51, 2012
|
|
2L44
 
 | | C-terminal zinc knuckle of the HIVNCp7 | | Descriptor: | C-terminal Zinc Knucle of the HIV-NCP7, ZINC ION | | Authors: | Quintal, S.M.O, Viegas, A.J.M, Cabrita, E.J, Farrell, N.P, Erhardt, S. | | Deposit date: | 2010-10-01 | | Release date: | 2011-10-12 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Platinated DNA affects zinc finger conformation. Interaction of a platinated single-stranded oligonucleotide and the C-terminal zinc finger of nucleocapsid protein HIVNCp7.
Biochemistry, 51, 2012
|
|
