4WVM
 
 | | Stonustoxin structure | | Descriptor: | Stonustoxin subunit alpha, Stonustoxin subunit beta | | Authors: | Ellisdon, A.M, Panjikar, S, Whisstock, J.C, McGowan, S. | | Deposit date: | 2014-11-06 | | Release date: | 2015-12-02 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Stonefish toxin defines an ancient branch of the perforin-like superfamily.
Proc.Natl.Acad.Sci.USA, 112, 2015
|
|
5HN1
 
 | | Crystal structure of Interleukin-37 | | Descriptor: | Interleukin-37, SULFATE ION | | Authors: | Ellisdon, A.M, Nold-Petry, C.A, Nold, M.F, Whisstock, J.C. | | Deposit date: | 2016-01-17 | | Release date: | 2017-02-22 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Homodimerization attenuates the anti-inflammatory activity of interleukin-37.
Sci Immunol, 2, 2017
|
|
7RX9
 
 | | Structure of autoinhibited P-Rex1 | | Descriptor: | Phosphatidylinositol 3,4,5-trisphosphate-dependent Rac exchanger 1 protein, Endolysin chimera, SULFATE ION | | Authors: | Ellisdon, A.M, Chang, Y. | | Deposit date: | 2021-08-22 | | Release date: | 2022-08-10 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (3.22 Å) | | Cite: | Structure of the metastatic factor P-Rex1 reveals a two-layered autoinhibitory mechanism.
Nat.Struct.Mol.Biol., 29, 2022
|
|
4P0O
 
 | | Cleaved Serpin 42Da | | Descriptor: | Serine protease inhibitor 4, isoform B | | Authors: | Ellisdon, A.M, Whisstock, J.C. | | Deposit date: | 2014-02-21 | | Release date: | 2014-05-07 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | High resolution structure of cleaved Serpin 42 Da from Drosophila melanogaster.
Bmc Struct.Biol., 14, 2014
|
|
4P0F
 
 | | Cleaved Serpin 42Da (C 2 2 21) | | Descriptor: | Serine protease inhibitor 4, isoform B | | Authors: | Ellisdon, A.M, Whisstock, J.C. | | Deposit date: | 2014-02-21 | | Release date: | 2014-05-07 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | High resolution structure of cleaved Serpin 42 Da from Drosophila melanogaster.
Bmc Struct.Biol., 14, 2014
|
|
4YON
 
 | | P-Rex1:Rac1 complex | | Descriptor: | Phosphatidylinositol 3,4,5-trisphosphate-dependent Rac exchanger 1 protein, Ras-related C3 botulinum toxin substrate 1 | | Authors: | Lucato, C.M, Whisstock, J.C, Ellisdon, A.M. | | Deposit date: | 2015-03-11 | | Release date: | 2015-07-01 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | The Phosphatidylinositol (3,4,5)-Trisphosphate-dependent Rac Exchanger 1Ras-related C3 Botulinum Toxin Substrate 1 (P-Rex1Rac1) Complex Reveals the Basis of Rac1 Activation in Breast Cancer Cells.
J.Biol.Chem., 290, 2015
|
|
7MOC
 
 | | Neurofibromin core | | Descriptor: | Isoform I of Neurofibromin | | Authors: | Lupton, C.J, Bayly-Jones, C, Ellisdon, A.M. | | Deposit date: | 2021-05-01 | | Release date: | 2021-12-15 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (4.56 Å) | | Cite: | The cryo-EM structure of the human neurofibromin dimer reveals the molecular basis for neurofibromatosis type 1.
Nat.Struct.Mol.Biol., 28, 2021
|
|
3KJL
 
 | | Sgf11:Sus1 complex | | Descriptor: | Protein SUS1, SAGA-associated factor 11 | | Authors: | Stewart, M, Ellisdon, A.M. | | Deposit date: | 2009-11-03 | | Release date: | 2009-12-08 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structural basis for the interaction between yeast Spt-Ada-Gcn5 acetyltransferase (SAGA) complex components Sgf11 and Sus1.
J.Biol.Chem., 285, 2010
|
|
3T5V
 
 | | Sac3:Thp1:Sem1 complex | | Descriptor: | 26S proteasome complex subunit SEM1, Nuclear mRNA export protein SAC3, Nuclear mRNA export protein THP1 | | Authors: | Stewart, M, Ellisdon, A.M. | | Deposit date: | 2011-07-28 | | Release date: | 2012-02-22 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structural basis for the assembly and nucleic acid binding of the TREX-2 transcription-export complex.
Nat.Struct.Mol.Biol., 19, 2012
|
|
3T5X
 
 | | PCID2:DSS1 Structure | | Descriptor: | 26S proteasome complex subunit DSS1, PCI domain-containing protein 2 | | Authors: | Stewart, M, Ellisdon, A.M. | | Deposit date: | 2011-07-28 | | Release date: | 2012-02-22 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.123 Å) | | Cite: | Structural basis for the assembly and nucleic acid binding of the TREX-2 transcription-export complex.
Nat.Struct.Mol.Biol., 19, 2012
|
|
2QP2
 
 | | Structure of a MACPF/perforin-like protein | | Descriptor: | CALCIUM ION, Unknown protein | | Authors: | Rosado, C.J, Buckle, A.M, Law, R.H.P, Butcher, R.E, Kan, W.T, Bird, C.H, Ung, K, Browne, K.A, Baran, K, Bashtannyk-Puhalovich, T.A, Faux, N.G, Wong, W, Porter, C.J, Pike, R.N, Ellisdon, A.M, Pearce, M.C, Bottomley, S.P, Emsley, J, Smith, A.I, Rossjohn, J, Hartland, E.L, Voskoboinik, I, Trapani, J.A, Bird, P.I, Dunstone, M.A, Whisstock, J.C. | | Deposit date: | 2007-07-22 | | Release date: | 2007-09-04 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | A common fold mediates vertebrate defense and bacterial attack
Science, 317, 2007
|
|
9C9I
 
 | | Structure of the TSC1:WIPI3 complex | | Descriptor: | Hamartin, WD repeat domain phosphoinositide-interacting protein 3 | | Authors: | D'Andrea, L, Bayly-Jones, C, Lupton, C.J, Ellisdon, A.M. | | Deposit date: | 2024-06-14 | | Release date: | 2024-12-04 | | Method: | X-RAY DIFFRACTION (3.18 Å) | | Cite: | Structure of the human TSC:WIPI3 lysosomal recruitment complex.
Sci Adv, 10, 2024
|
|
9CE3
 
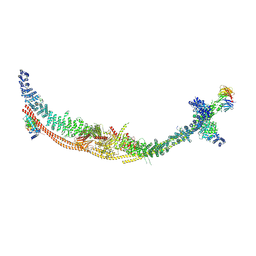 | | Structure of the TSC:WIPI3 lysosomal recruitment complex | | Descriptor: | Hamartin, Isoform 4 of Tuberin, TBC1 domain family member 7, ... | | Authors: | Bayly-Jones, C, Lupton, C.J, D'Andrea, L, Ellisdon, A.M. | | Deposit date: | 2024-06-26 | | Release date: | 2024-12-04 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Structure of the human TSC:WIPI3 lysosomal recruitment complex.
Sci Adv, 10, 2024
|
|
7SYF
 
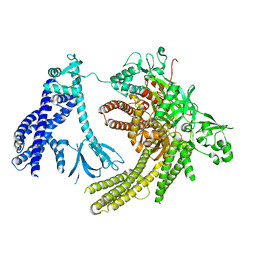 | | Reconstruction of full-length Prex-1 (PtdIns(3,4,5)P3-dependent Rac Exchanger 1) | | Descriptor: | Phosphatidylinositol 3,4,5-trisphosphate-dependent Rac exchanger 1 protein,Endolysin chimera | | Authors: | Lupton, C.J, Bayly-Jones, C, Ellisdon, A.M. | | Deposit date: | 2021-11-24 | | Release date: | 2022-07-27 | | Last modified: | 2024-02-28 | | Method: | ELECTRON MICROSCOPY (4.2 Å) | | Cite: | Structure of the metastatic factor P-Rex1 reveals a two-layered autoinhibitory mechanism.
Nat.Struct.Mol.Biol., 29, 2022
|
|
7MP5
 
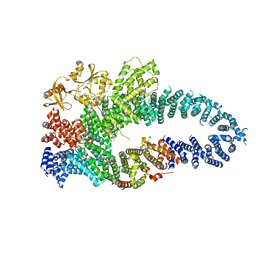 | | Autoinhibited neurofibrobmin | | Descriptor: | Isoform I of Neurofibromin | | Authors: | Lupton, C.J, Bayly-Jones, C, Ellisdon, A.M. | | Deposit date: | 2021-05-04 | | Release date: | 2021-12-15 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (5.6 Å) | | Cite: | The cryo-EM structure of the human neurofibromin dimer reveals the molecular basis for neurofibromatosis type 1.
Nat.Struct.Mol.Biol., 28, 2021
|
|
7MP6
 
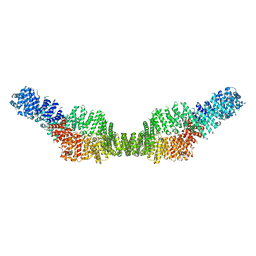 | | Neurofibromin homodimer | | Descriptor: | Isoform I of Neurofibromin | | Authors: | Lupton, C.J, Bayly-Jones, C, Ellisdon, A.M. | | Deposit date: | 2021-05-04 | | Release date: | 2021-12-15 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (6.25 Å) | | Cite: | The cryo-EM structure of the human neurofibromin dimer reveals the molecular basis for neurofibromatosis type 1.
Nat.Struct.Mol.Biol., 28, 2021
|
|
3KIK
 
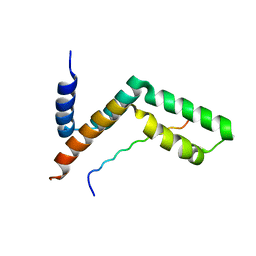 | | Sgf11:Sus1 complex | | Descriptor: | Protein SUS1, SAGA-associated factor 11 | | Authors: | Stewart, M, Ellisdon, A.M. | | Deposit date: | 2009-11-02 | | Release date: | 2009-12-08 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural basis for the interaction between yeast Spt-Ada-Gcn5 acetyltransferase (SAGA) complex components Sgf11 and Sus1
J.Biol.Chem., 285, 2010
|
|
8U5V
 
 | | The mTORC1 cholesterol sensor LYCHOS (GPR155) - auxin bound, closed state | | Descriptor: | 1H-INDOL-3-YLACETIC ACID, CHOLESTEROL HEMISUCCINATE, Integral membrane protein GPR155 | | Authors: | Bayly-Jones, C, Lupton, C.J, Ellisdon, A.M. | | Deposit date: | 2023-09-13 | | Release date: | 2024-09-18 | | Last modified: | 2024-11-13 | | Method: | ELECTRON MICROSCOPY (2.77 Å) | | Cite: | LYCHOS is a human hybrid of a plant-like PIN transporter and a GPCR.
Nature, 634, 2024
|
|
8U54
 
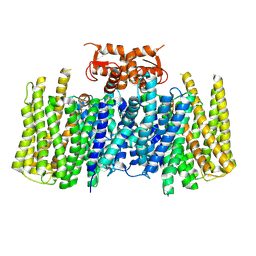 | |
8U58
 
 | |
8U5X
 
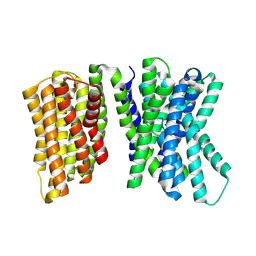 | |
8U5Q
 
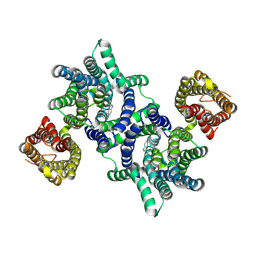 | |
8U56
 
 | |
8U5C
 
 | |
8U5N
 
 | |
