2VA5
 
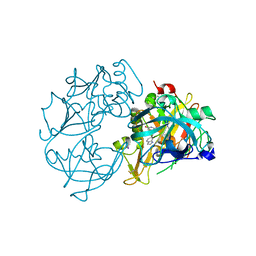 | | X-ray crystal structure of beta secretase complexed with compound 8c | | Descriptor: | 2-amino-6-[2-(1H-indol-6-yl)ethyl]pyrimidin-4(3H)-one, BETA-SECRETASE 1 ., IODIDE ION | | Authors: | Edwards, P.D, Albert, J.S, Sylvester, M, Aharony, D, Andisik, D, Callaghan, O, Campbell, J.B, Carr, R.A, Chessari, G, Congreve, M, Frederickson, M, Folmer, R.H.A, Geschwindner, S, Koether, G, Kolmodin, K, Krumrine, J, Mauger, R.C, Murray, C.W, Olsson, L, Patel, S, Spear, N, Tian, G. | | Deposit date: | 2007-08-30 | | Release date: | 2007-11-13 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Application of Fragment-Based Lead Generation to the Discovery of Novel, Cyclic Amidine Beta-Secretase Inhibitors with Nanomolar Potency, Cellular Activity, and High Ligand Efficiency.
J.Med.Chem., 50, 2007
|
|
2VA7
 
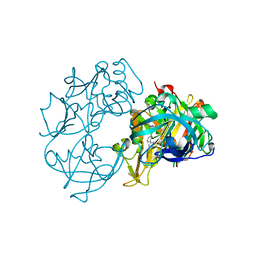 | | X-ray crystal structure of beta secretase complexed with compound 27 | | Descriptor: | (6R)-2-amino-6-[2-(3'-methoxybiphenyl-3-yl)ethyl]-3,6-dimethyl-5,6-dihydropyrimidin-4(3H)-one, BETA-SECRETASE 1 ., IODIDE ION | | Authors: | Edwards, P.D, Albert, J.S, Sylvester, M, Aharony, D, Andisik, D, Callaghan, O, Campbell, J.B, Carr, R.A, Chessari, G, Congreve, M, Frederickson, M, Folmer, R.H.A, Geschwindner, S, Koether, G, Kolmodin, K, Krumrine, J, Mauger, R.C, Murray, C.W, Olsson, L.L, Patel, S, Spear, N, Tian, G. | | Deposit date: | 2007-08-30 | | Release date: | 2007-11-13 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Application of Fragment-Based Lead Generation to the Discovery of Novel, Cyclic Amidine Beta-Secretase Inhibitors with Nanomolar Potency, Cellular Activity, and High Ligand Efficiency.
J.Med.Chem., 50, 2007
|
|
2VA6
 
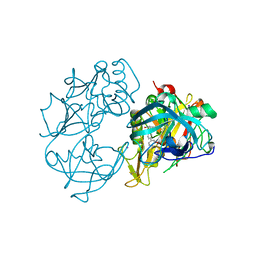 | | X-ray crystal structure of beta secretase complexed with compound 24 | | Descriptor: | (6S)-2-amino-6-(3'-methoxybiphenyl-3-yl)-3,6-dimethyl-5,6-dihydropyrimidin-4(3H)-one, BETA SECRETASE 1, IODIDE ION | | Authors: | Edwards, P.D, Albert, J.S, Sylvester, M, Aharony, D, Andisik, D, Callaghan, O, Campbell, J.B, Carr, R.A, Chessari, G, Congreve, M, Frederickson, M, Folmer, R.H.A, Geschwindner, S, Koether, G, Kolmodin, K, Krumrine, J, Mauger, R.C, Murray, C.W, Olsson, L.L, Patel, S, Spear, N, Tian, G. | | Deposit date: | 2007-08-30 | | Release date: | 2007-11-13 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Application of Fragment-Based Lead Generation to the Discovery of Novel, Cyclic Amidine Beta-Secretase Inhibitors with Nanomolar Potency, Cellular Activity, and High Ligand Efficiency.
J.Med.Chem., 50, 2007
|
|
1E5M
 
 | |
8PM2
 
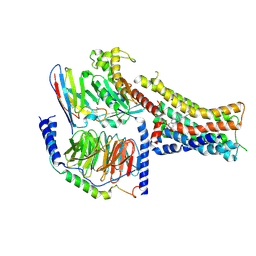 | | Structure of the murine trace amine-associated receptor TAAR7f bound to N,N-dimethylcyclohexylamine (DMCH) in complex with mini-Gs trimeric G protein | | Descriptor: | CHOLESTEROL HEMISUCCINATE, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | Authors: | Gusach, A, Lee, Y, Edwards, P.C, Huang, F, Weyand, S.N, Tate, C.G. | | Deposit date: | 2023-06-28 | | Release date: | 2023-08-09 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (2.92 Å) | | Cite: | Molecular recognition of an aversive odorant by the murine trace amine-associated receptor TAAR7f.
Biorxiv, 2023
|
|
6H7L
 
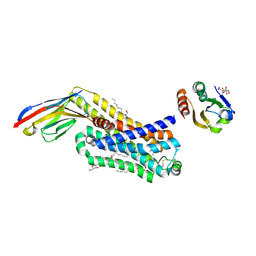 | | ACTIVATED TURKEY BETA1 ADRENOCEPTOR WITH BOUND PARTIAL AGONIST DOBUTAMINE AND NANOBODY Nb6B9 | | Descriptor: | Beta-1 adrenergic receptor, Camelid antibody fragment Nb6B9, DOBUTAMINE, ... | | Authors: | Warne, T, Edwards, P.C, Dore, A.S, Leslie, A.G.W, Tate, C.G. | | Deposit date: | 2018-07-31 | | Release date: | 2018-10-17 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Molecular basis for high-affinity agonist binding in GPCRs.
Science, 364, 2019
|
|
6H7O
 
 | | ACTIVATED TURKEY BETA1 ADRENOCEPTOR WITH BOUND WEAK PARTIAL AGONIST CYANOPINDOLOL AND NANOBODY Nb6B9 | | Descriptor: | 4-{[(2S)-3-(tert-butylamino)-2-hydroxypropyl]oxy}-3H-indole-2-carbonitrile, Beta-1 adrenergic receptor, Camelid antibody fragment Nb6B9, ... | | Authors: | Warne, T, Edwards, P.C, Dore, A.S, Leslie, A.G.W, Tate, C.G. | | Deposit date: | 2018-07-31 | | Release date: | 2018-10-17 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Molecular basis for high-affinity agonist binding in GPCRs.
Science, 364, 2019
|
|
6H7N
 
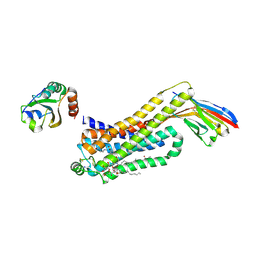 | | ACTIVATED TURKEY BETA1 ADRENOCEPTOR WITH BOUND PARTIAL AGONIST XAMOTEROL AND NANOBODY Nb6B9 | | Descriptor: | Beta-1 adrenergic receptor, Camelid antibody fragment Nb6B9, HEGA-10, ... | | Authors: | Warne, T, Edwards, P.C, Dore, A.S, Leslie, A.G.W, Tate, C.G. | | Deposit date: | 2018-07-31 | | Release date: | 2018-10-17 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Molecular basis for high affinity agonist binding in GPCRs
Biorxiv, 2018
|
|
6H7M
 
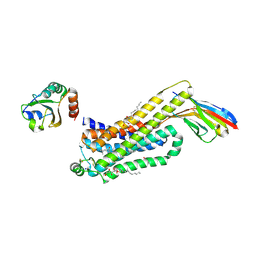 | | ACTIVATED TURKEY BETA1 ADRENOCEPTOR WITH BOUND PARTIAL AGONIST SALBUTAMOL AND NANOBODY Nb6B9 | | Descriptor: | Beta-1 adrenergic receptor, Camelid antibody fragment Nb6B9, HEGA-10, ... | | Authors: | Warne, T, Edwards, P.C, Dore, A.S, Leslie, A.G.W, Tate, C.G. | | Deposit date: | 2018-07-31 | | Release date: | 2018-10-17 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.76 Å) | | Cite: | Molecular basis for high-affinity agonist binding in GPCRs.
Science, 364, 2019
|
|
8PWR
 
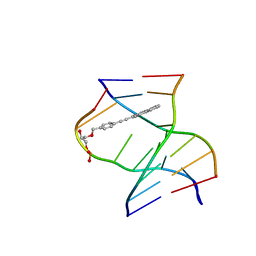 | | TINA-conjugated antiparallel DNA triplex | | Descriptor: | DNA (5'-D(*AP*GP*GP*AP*GP*GP*A)-3'), DNA (5'-D(*TP*CP*CP*TP*CP*CP*T)-3'), DNA (5'-D(*TP*GP*GP*TP*GP*(J32)P*GP*T)-3') | | Authors: | Garavis, M, Edwards, P.J.B, Serrano-Chacon, I, Doluca, O, Filichev, V.V, Gonzalez, C. | | Deposit date: | 2023-07-21 | | Release date: | 2024-01-17 | | Last modified: | 2024-03-27 | | Method: | SOLUTION NMR | | Cite: | Understanding intercalative modulation of G-rich sequence folding: solution structure of a TINA-conjugated antiparallel DNA triplex.
Nucleic Acids Res., 52, 2024
|
|
4A4M
 
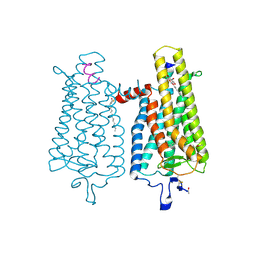 | | Crystal structure of the light-activated constitutively active N2C, M257Y,D282C rhodopsin mutant in complex with a peptide resembling the C-terminus of the Galpha-protein subunit (GaCT) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ACETATE ION, GUANINE NUCLEOTIDE-BINDING PROTEIN G(T) SUBUNIT ALPHA-3, ... | | Authors: | Deupi, X, Edwards, P, Singhal, A, Nickle, B, Oprian, D.D, Schertler, G.F.X, Standfuss, J. | | Deposit date: | 2011-10-17 | | Release date: | 2012-01-25 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Stabilized G Protein Binding Site in the Structure of Constitutively Active Metarhodopsin-II.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
6U1L
 
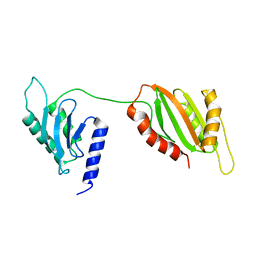 | | Structure of two-domain translational regulator Yih1 reveals a possible mechanism of action | | Descriptor: | Protein IMPACT homolog | | Authors: | Harjes, E, Jameson, G.B, Edwards, P.J.B, Goroncy, A.K, Loo, T, Norris, G.E. | | Deposit date: | 2019-08-16 | | Release date: | 2021-02-10 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Experimentally based structural model of Yih1 provides insight into its function in controlling the key translational regulator Gcn2.
Febs Lett., 595, 2021
|
|
6U1O
 
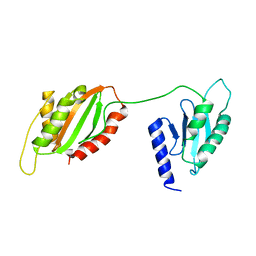 | | Structure of two-domain translational regulator Yih1 reveals a possible mechanism of action | | Descriptor: | Protein IMPACT homolog | | Authors: | Harjes, E, Jameson, G.B, Edwards, P.J.B, Goroncy, A.K, Loo, T, Norris, G.E. | | Deposit date: | 2019-08-16 | | Release date: | 2021-02-10 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Experimentally based structural model of Yih1 provides insight into its function in controlling the key translational regulator Gcn2.
Febs Lett., 595, 2021
|
|
4UG2
 
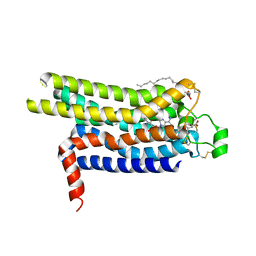 | | Thermostabilised HUMAN A2a Receptor with CGS21680 bound | | Descriptor: | (2S)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, 2-[P-(2-CARBOXYETHYL)PHENYLETHYL-AMINO]-5'-N-ETHYLCARBOXAMIDO ADENOSINE, THERMOSTABILISED HUMAN A2A RECEPTOR | | Authors: | Lebon, G, Edwards, P.C, Leslie, A.G.W, Tate, C.G. | | Deposit date: | 2015-03-21 | | Release date: | 2015-04-08 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Molecular Determinants of Cgs21680 Binding to the Human Adenosine A2A Receptor.
Mol.Pharmacol., 87, 2015
|
|
4UHR
 
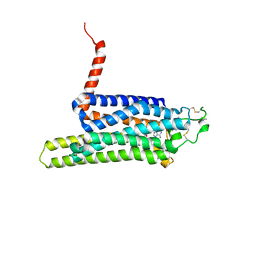 | | Thermostabilised HUMAN A2a Receptor with CGS21680 bound | | Descriptor: | 2-[P-(2-CARBOXYETHYL)PHENYLETHYL-AMINO]-5'-N-ETHYLCARBOXAMIDO ADENOSINE, THERMOSTABILISED HUMAN A2A RECEPTOR | | Authors: | Lebon, G, Edwards, P.C, Leslie, A.G.W, Tate, C.G. | | Deposit date: | 2015-03-25 | | Release date: | 2015-04-08 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Molecular Determinants of Cgs21680 Binding to the Human Adenosine A2A Receptor.
Mol.Pharmacol., 87, 2015
|
|
6IBL
 
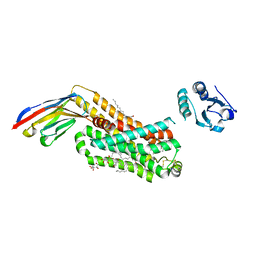 | | ACTIVATED TURKEY BETA1 ADRENOCEPTOR WITH BOUND AGONIST FORMOTEROL AND NANOBODY Nb80 | | Descriptor: | Camelid antibody fragment Nb80, HEGA-10, SODIUM ION, ... | | Authors: | Warne, T, Edwards, P.C, Dore, A.S, Leslie, A.G.W, Tate, C.G. | | Deposit date: | 2018-11-30 | | Release date: | 2019-01-09 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Molecular basis of beta-arrestin coupling to formoterol-bound beta1-adrenoceptor.
Nature, 583, 2020
|
|
2KUY
 
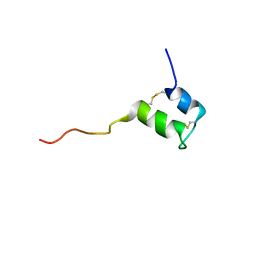 | | Structure of Glycocin F | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Prebacteriocin glycocin F | | Authors: | Venugopal, H, Edwards, P, Schwalbe, M, Claridge, J, Stepper, J, Patchett, M, Loo, T, Libich, D, Norris, G, Pascal, S. | | Deposit date: | 2010-03-01 | | Release date: | 2011-05-04 | | Last modified: | 2024-11-20 | | Method: | SOLUTION NMR | | Cite: | Structural, dynamic, and chemical characterization of a novel s-glycosylated bacteriocin.
Biochemistry, 50, 2011
|
|
8DFZ
 
 | |
4AMJ
 
 | | Turkey beta1 adrenergic receptor with stabilising mutations and bound biased agonist carvedilol | | Descriptor: | (2S)-1-(8H-CARBAZOL-4-YLOXY)-3-[2-(2-METHOXYPHENOXY)ETHYLAMINO]PROPAN-2-OL, BETA-1 ADRENERGIC RECEPTOR, HEGA-10, ... | | Authors: | Warne, T, Edwards, P.C, Leslie, A.G, Tate, C.G. | | Deposit date: | 2012-03-12 | | Release date: | 2012-05-23 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal Structures of a Stabilized Beta1-Adrenoceptor Bound to the Biased Agonists Bucindolol and Carvedilol
Structure, 20, 2012
|
|
4AMI
 
 | | Turkey beta1 adrenergic receptor with stabilising mutations and bound biased agonist bucindolol | | Descriptor: | 2-[(2S)-3-[[1-(1H-indol-3-yl)-2-methyl-propan-2-yl]amino]-2-oxidanyl-propoxy]benzenecarbonitrile, BETA-1 ADRENERGIC RECEPTOR, HEGA-10 | | Authors: | Warne, T, Edwards, P.C, Leslie, A.G, Tate, C.G. | | Deposit date: | 2012-03-11 | | Release date: | 2012-05-23 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Crystal Structures of a Stabilized Beta1-Adrenoceptor Bound to the Biased Agonists Bucindolol and Carvedilol
Structure, 20, 2012
|
|
4BVN
 
 | | Ultra-thermostable beta1-adrenoceptor with cyanopindolol bound | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, (CARBAMOYLMETHYL-CARBOXYMETHYL-AMINO)-ACETIC ACID, 4-{[(2S)-3-(tert-butylamino)-2-hydroxypropyl]oxy}-3H-indole-2-carbonitrile, ... | | Authors: | Miller, J, Nehme, R, Warne, T, Edwards, P.C, Leslie, A.G.W, Schertler, G, Tate, C.G. | | Deposit date: | 2013-06-26 | | Release date: | 2014-04-02 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | The 2.1 A Resolution Structure of Cyanopindolol-Bound Beta1- Adrenoceptor Identifies an Intramembrane Na+ Ion that Stabilises the Ligand-Free Receptor.
Plos One, 9, 2014
|
|
6BQI
 
 | | Structure of two-domain translational regulator Yih1 reveals a possible mechanism of action | | Descriptor: | Protein IMPACT homolog | | Authors: | Harjes, E, Jameson, G.B, Edwards, P.J.B, Goroncy, A.K, Loo, T, Norris, G.E. | | Deposit date: | 2017-11-27 | | Release date: | 2018-11-28 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR, SOLUTION SCATTERING | | Cite: | Experimentally based structural model of Yih1 provides insight into its function in controlling the key translational regulator Gcn2.
Febs Lett., 2020
|
|
2JIC
 
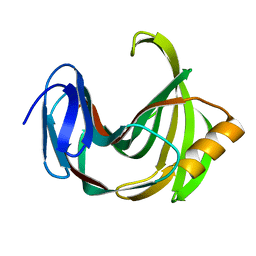 | | High resolution structure of xylanase-II from one micron beam experiment | | Descriptor: | XYLANASE-II | | Authors: | Moukhametzianov, R, Burghammer, M, Edwards, P.C, Petitdemange, S, Popov, D, Fransen, M, Schertler, G.F, Riekel, C. | | Deposit date: | 2007-02-27 | | Release date: | 2008-05-13 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Protein Crystallography with a Micrometre-Sized Synchrotron-Radiation Beam.
Acta Crystallogr.,Sect.D, 64, 2008
|
|
1OSH
 
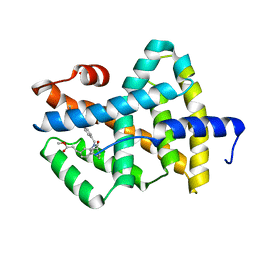 | | A Chemical, Genetic, and Structural Analysis of the nuclear bile acid receptor FXR | | Descriptor: | Bile acid receptor, METHYL 3-{3-[(CYCLOHEXYLCARBONYL){[4'-(DIMETHYLAMINO)BIPHENYL-4-YL]METHYL}AMINO]PHENYL}ACRYLATE | | Authors: | Downes, M, Verdecia, M.A, Roecker, A.J, Hughes, R, Hogenesch, J.B, Kast-Woelbern, H.R, Bowman, M.E, Ferrer, J.-L, Anisfeld, A.M, Edwards, P.A, Rosenfeld, J.M, Alvarez, J.G.A, Noel, J.P, Nicolaou, K.C, Evans, R.M. | | Deposit date: | 2003-03-19 | | Release date: | 2003-09-23 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | A chemical, genetic, and structural analysis of the nuclear bile acid receptor FXR
Mol.Cell, 11, 2003
|
|
2KXG
 
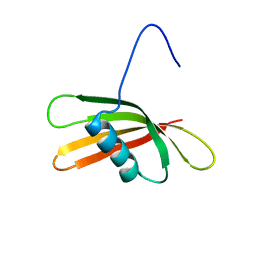 | | The solution structure of the squash aspartic acid proteinase inhibitor (SQAPI) | | Descriptor: | Aspartic protease inhibitor | | Authors: | Headey, S.J, Macaskill, U.K, Wright, M, Claridge, J.K, Edwards, P.J.B, Farley, P.C, Christeller, J.T, Laing, W.A, Pascal, S.M. | | Deposit date: | 2010-05-05 | | Release date: | 2010-06-09 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the squash aspartic acid proteinase inhibitor (SQAPI) and mutational analysis of pepsin inhibition.
J.Biol.Chem., 285, 2010
|
|
