6NJP
 
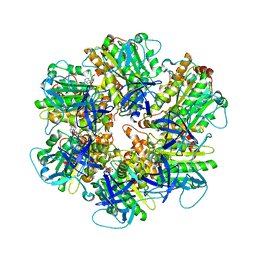 | | Structure of the assembled ATPase EscN in complex with its central stalk EscO from the enteropathogenic E. coli (EPEC) type III secretion system | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ALUMINUM FLUORIDE, EscO, ... | | Authors: | Majewski, D.D, Worrall, L.J, Hong, C, Atkinson, C.E, Vuckovic, M, Watanabe, N, Yu, Z, Strynadka, N.C.J. | | Deposit date: | 2019-01-03 | | Release date: | 2019-02-20 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (3.29 Å) | | Cite: | Cryo-EM structure of the homohexameric T3SS ATPase-central stalk complex reveals rotary ATPase-like asymmetry.
Nat Commun, 10, 2019
|
|
7RYG
 
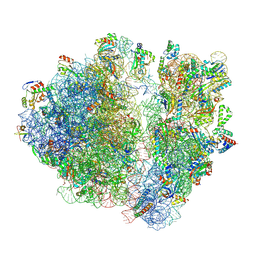 | | A. baumannii Ribosome-TP-6076 complex: E-site tRNA 70S | | Descriptor: | (4S,4aS,5aR,12aS)-4-(diethylamino)-3,10,12,12a-tetrahydroxy-1,11-dioxo-8-[(2S)-pyrrolidin-2-yl]-7-(trifluoromethyl)-1,4,4a,5,5a,6,11,12a-octahydrotetracene-2-carboxamide, 16S Ribosomal RNA, 23S ribosomal RNA, ... | | Authors: | Morgan, C.E, Yu, E.W. | | Deposit date: | 2021-08-25 | | Release date: | 2022-02-02 | | Last modified: | 2024-06-05 | | Method: | ELECTRON MICROSCOPY (2.38 Å) | | Cite: | An Analysis of the Novel Fluorocycline TP-6076 Bound to Both the Ribosome and Multidrug Efflux Pump AdeJ from Acinetobacter baumannii.
Mbio, 13, 2022
|
|
5N99
 
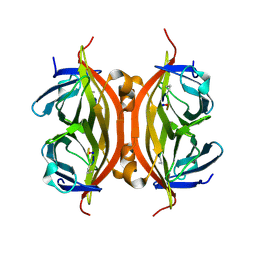 | | CRYSTAL STRUCTURE OF STREPTAVIDIN with cyclic peptide NQpWQ | | Descriptor: | ASN-GLN-DPR-TRP-GLN, Streptavidin | | Authors: | Lyamichev, V, Goodrich, L, Sullivan, E, Bannen, R, Benz, J, Albert, T, Patel, J. | | Deposit date: | 2017-02-24 | | Release date: | 2017-10-04 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Stepwise Evolution Improves Identification of Diverse Peptides Binding to a Protein Target.
Sci Rep, 7, 2017
|
|
7OB1
 
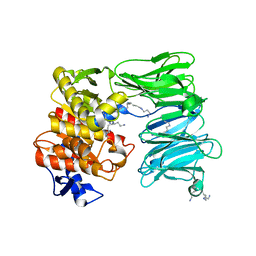 | | OLIGOPEPTIDASE B FROM S. PROTEOMACULANS WITH MODIFIED HINGE | | Descriptor: | Oligopeptidase B, SPERMINE | | Authors: | Petrenko, D.E, Nikolaeva, A.Y, Lazarenko, V.A, Dorovatovskiy, P.V, Vlaskina, A.V, Korzhenevskiy, D.A, Mikhailova, A.G, Rakitina, T.V, Timofeev, V.I. | | Deposit date: | 2021-04-20 | | Release date: | 2021-05-19 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | First Crystal Structure of Bacterial Oligopeptidase B in an Intermediate State: The Roles of the Hinge Region Modification and Spermine.
Biology (Basel), 10, 2021
|
|
8K65
 
 | | Serial femtosecond crystallography structure of CO bound ba3- type cytochrome c oxidase without pump laser irradiation | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, CARBON MONOXIDE, COPPER (II) ION, ... | | Authors: | Safari, C, Ghosh, S, Andersson, R, Johannesson, J, Donoso, A.V, Bath, P, Zoric, D, Sandelin, E, Nango, E, Tanaka, R, Iwata, S, Neutze, R, Branden, G. | | Deposit date: | 2023-07-25 | | Release date: | 2023-11-15 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Time-resolved serial crystallography to track the dynamics of carbon monoxide in the active site of cytochrome c oxidase.
Sci Adv, 9, 2023
|
|
6N64
 
 | | Crystal structure of mouse SMCHD1 hinge domain | | Descriptor: | Structural maintenance of chromosomes flexible hinge domain-containing protein 1, Uncharacterized peptide from Structural maintenance of chromosomes flexible hinge domain-containing protein 1 | | Authors: | Birkinshaw, R.W, Chen, K, Czabotar, P.E, Blewitt, M.E, Murphy, J.M. | | Deposit date: | 2018-11-25 | | Release date: | 2020-06-17 | | Last modified: | 2021-01-20 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Crystal structure of the hinge domain of Smchd1 reveals its dimerization mode and nucleic acid-binding residues.
Sci.Signal., 13, 2020
|
|
5EGO
 
 | | HOXB13-MEIS1 heterodimer bound to methylated DNA | | Descriptor: | DNA (5'-D(*CP*CP*TP*(5CM)P*GP*TP*AP*AP*AP*AP*CP*TP*GP*TP*CP*AP*AP*C)-3'), DNA (5'-D(P*GP*TP*TP*GP*AP*CP*AP*GP*TP*TP*TP*TP*AP*(5CM)P*GP*AP*GP*G)-3'), Homeobox protein Hox-B13, ... | | Authors: | Morgunova, E, Yin, Y, Jolma, A, Popov, A, Taipale, J. | | Deposit date: | 2015-10-27 | | Release date: | 2016-11-09 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.54 Å) | | Cite: | Impact of cytosine methylation on DNA binding specificities of human transcription factors.
Science, 356, 2017
|
|
7ON9
 
 | | Crystal structure of para-hydroxybenzoate-3-hydroxylase PraI | | Descriptor: | 4-hydroxybenzoate 3-monooxygenase (NAD(P)H), FLAVIN-ADENINE DINUCLEOTIDE, P-HYDROXYBENZOIC ACID | | Authors: | Zahn, M, McGeehan, J.E. | | Deposit date: | 2021-05-25 | | Release date: | 2022-01-26 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.63 Å) | | Cite: | Debottlenecking 4-hydroxybenzoate hydroxylation in Pseudomonas putida KT2440 improves muconate productivity from p-coumarate.
Metab Eng, 70, 2022
|
|
8SUF
 
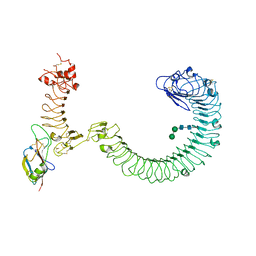 | | The complex of TOL-1 ectodomain bound to LAT-1 Lectin domain | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Latrophilin-like protein 1, ... | | Authors: | Carmona Rosas, G, Li, J, Arac, D, Ozkan, E. | | Deposit date: | 2023-05-12 | | Release date: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (4 Å) | | Cite: | Structural basis and functional roles for Toll-like receptor binding to Latrophilin adhesion-GPCR in embryo development
To Be Published
|
|
6PT0
 
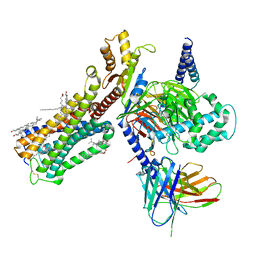 | | Cryo-EM structure of human cannabinoid receptor 2-Gi protein in complex with agonist WIN 55,212-2 | | Descriptor: | CHOLESTEROL, Cannabinoid receptor 2, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, ... | | Authors: | Xu, T.H, Xing, C, Zhuang, Y, Feng, Z, Zhou, X.E, Chen, M, Wang, L, Meng, X, Xue, Y, Wang, J, Liu, H, McGuire, T, Zhao, G, Melcher, K, Zhang, C, Xu, H.E, Xie, X.Q. | | Deposit date: | 2019-07-14 | | Release date: | 2020-02-12 | | Last modified: | 2020-03-04 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Cryo-EM Structure of the Human Cannabinoid Receptor CB2-GiSignaling Complex.
Cell, 180, 2020
|
|
6SSI
 
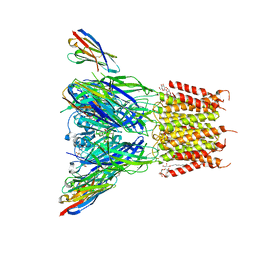 | | Structure of the pentameric ligand-gated ion channel ELIC in complex with a PAM nanobody | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, ACETATE ION, CALCIUM ION, ... | | Authors: | Ulens, C, Brams, M, Evans, G.L, Spurny, R, Govaerts, C, Pardon, E, Steyaert, J. | | Deposit date: | 2019-09-07 | | Release date: | 2020-02-12 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.59 Å) | | Cite: | Modulation of the Erwinia ligand-gated ion channel (ELIC) and the 5-HT 3 receptor via a common vestibule site.
Elife, 9, 2020
|
|
5EF6
 
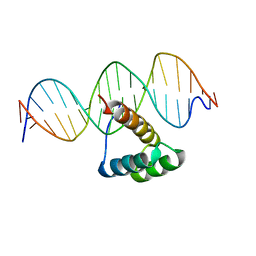 | | Structure of HOXB13 complex with methylated DNA | | Descriptor: | DNA (5'-D(P*GP*GP*AP*CP*CP*TP*(5CM)P*GP*TP*AP*AP*AP*AP*CP*AP*CP*AP*A)-3'), DNA (5'-D(P*TP*TP*GP*TP*GP*TP*TP*TP*TP*AP*(5CM)P*GP*AP*GP*GP*TP*CP*C)-3'), Homeobox protein Hox-B13 | | Authors: | Morgunova, E, Yin, Y, Jolma, A, Popov, A, Taipale, J. | | Deposit date: | 2015-10-23 | | Release date: | 2017-02-08 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Impact of cytosine methylation on DNA binding specificities of human transcription factors.
Science, 356, 2017
|
|
8QO4
 
 | | Conserved Structures and Dynamics in 5-Proximal Regions of Betacoronavirus RNA Genomes | | Descriptor: | MERS-CoV-SL5 | | Authors: | Moura, T.R, Purta, E, Bernat, A, Baulin, E, Mukherjee, S, Bujnicki, J.M. | | Deposit date: | 2023-09-28 | | Release date: | 2024-03-06 | | Last modified: | 2024-04-24 | | Method: | ELECTRON MICROSCOPY (5.9 Å) | | Cite: | Conserved structures and dynamics in 5'-proximal regions of Betacoronavirus RNA genomes.
Nucleic Acids Res., 52, 2024
|
|
8OFE
 
 | | E.coli Peptide Deformylase with bound inhibitor | | Descriptor: | (2R)-2-[[methanoyl(oxidanyl)amino]methyl]-N-[(2S)-3-methyl-1-oxidanylidene-1-[2-(sulfanylmethyl)pyrrolidin-1-yl]butan-2-yl]heptanamide, Peptide deformylase, ZINC ION | | Authors: | Kirschner, H, Stoll, R, Hofmann, E. | | Deposit date: | 2023-03-15 | | Release date: | 2023-12-27 | | Last modified: | 2024-04-10 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Structural Insights into Antibacterial Payload Release from Gold Nanoparticles Bound to E. coli Peptide Deformylase.
Chemmedchem, 19, 2024
|
|
6Q4K
 
 | | CDK2 in complex with FragLite38 | | Descriptor: | (~{E})-3-[3-[(4-chlorophenyl)carbamoyl]phenyl]prop-2-enoic acid, 1,2-ETHANEDIOL, Cyclin-dependent kinase 2 | | Authors: | Wood, D.J, Martin, M.P, Noble, M.E.M. | | Deposit date: | 2018-12-05 | | Release date: | 2019-03-20 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.06 Å) | | Cite: | FragLites-Minimal, Halogenated Fragments Displaying Pharmacophore Doublets. An Efficient Approach to Druggability Assessment and Hit Generation.
J.Med.Chem., 62, 2019
|
|
8QO2
 
 | | Conserved Structures and Dynamics in 5-Proximal Regions of Betacoronavirus RNA Genomes | | Descriptor: | OC43-CoV-SL5 | | Authors: | Moura, T.R, Purta, E, Bernat, A, Baulin, E, Mukherjee, S, Bujnicki, J.M. | | Deposit date: | 2023-09-27 | | Release date: | 2024-03-06 | | Last modified: | 2024-04-24 | | Method: | ELECTRON MICROSCOPY (7.1 Å) | | Cite: | Conserved structures and dynamics in 5'-proximal regions of Betacoronavirus RNA genomes.
Nucleic Acids Res., 52, 2024
|
|
8QO3
 
 | | Conserved Structures and Dynamics in 5-Proximal Regions of Betacoronavirus RNA Genomes | | Descriptor: | RoBat-CoV-SL5 | | Authors: | Moura, T.R, Purta, E, Bernat, A, Baulin, E, Mukherjee, S, Bujnicki, J.M. | | Deposit date: | 2023-09-28 | | Release date: | 2024-03-06 | | Last modified: | 2024-04-24 | | Method: | ELECTRON MICROSCOPY (6.6 Å) | | Cite: | Conserved structures and dynamics in 5'-proximal regions of Betacoronavirus RNA genomes.
Nucleic Acids Res., 52, 2024
|
|
8QO5
 
 | | Conserved Structures and Dynamics in 5-Proximal Regions of Betacoronavirus RNA Genomes | | Descriptor: | SARS-CoV-2-SL5 | | Authors: | Moura, T.R, Purta, E, Bernat, A, Baulin, E, Mukherjee, S, Bujnicki, J.M. | | Deposit date: | 2023-09-28 | | Release date: | 2024-03-06 | | Last modified: | 2024-04-24 | | Method: | ELECTRON MICROSCOPY (6.5 Å) | | Cite: | Conserved structures and dynamics in 5'-proximal regions of Betacoronavirus RNA genomes.
Nucleic Acids Res., 52, 2024
|
|
6NBK
 
 | | Crystal structure of Arginase from Bacillus cereus | | Descriptor: | Arginase, CALCIUM ION, MANGANESE (II) ION | | Authors: | Chang, C, Evdokimova, E, Mcchesney, M, Joachimiak, A, Savchenko, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2018-12-07 | | Release date: | 2018-12-19 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.91 Å) | | Cite: | Crystal structure of Arginase from Bacillus cereus
To Be Published
|
|
4YUM
 
 | | Multiconformer synchrotron model of CypA at 300 K | | Descriptor: | Peptidyl-prolyl cis-trans isomerase A | | Authors: | Keedy, D.A, Kenner, L.R, Warkentin, M, Woldeyes, R.A, Thompson, M.C, Brewster, A.S, Van Benschoten, A.H, Baxter, E.L, Hopkins, J.B, Uervirojnangkoorn, M, McPhillips, S.E, Song, J, Mori, R.A, Holton, J.M, Weis, W.I, Brunger, A.T, Soltis, M, Lemke, H, Gonzalez, A, Sauter, N.K, Cohen, A.E, van den Bedem, H, Thorne, R.E, Fraser, J.S. | | Deposit date: | 2015-03-18 | | Release date: | 2015-10-14 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Mapping the conformational landscape of a dynamic enzyme by multitemperature and XFEL crystallography.
Elife, 4, 2015
|
|
5LN3
 
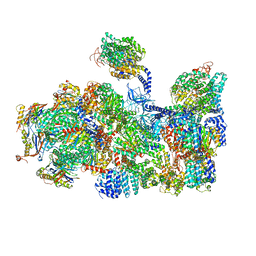 | | The human 26S Proteasome at 6.8 Ang. | | Descriptor: | 26S protease regulatory subunit 10B, 26S protease regulatory subunit 4, 26S protease regulatory subunit 6A, ... | | Authors: | Schweitzer, A, Beck, F, Sakata, E, Unverdorben, P. | | Deposit date: | 2016-08-03 | | Release date: | 2017-03-22 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (6.8 Å) | | Cite: | Molecular Details Underlying Dynamic Structures and Regulation of the Human 26S Proteasome.
Mol. Cell Proteomics, 16, 2017
|
|
6QJS
 
 | | Crystal structure of a DNA dodecamer containing a tetramethylpiperidinoxyl (nitroxide) spin label | | Descriptor: | COBALT HEXAMMINE(III), DNA (5'-D(P*GP*CP*AP*AP*AP*TP*TP*(S6M)P*GP*CP*G)-3') | | Authors: | Hardwick, J.S, Haugland, M.M, Ptchelkine, D, Brown, T, Anderson, E.E. | | Deposit date: | 2019-01-24 | | Release date: | 2020-02-12 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | 2'-Alkynyl spin-labelling is a minimally perturbing tool for DNA structural analysis.
Nucleic Acids Res., 48, 2020
|
|
6Z9W
 
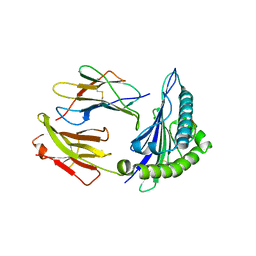 | | Human Class I Major Histocompatibility Complex, A02 allele, presenting LLGWVFAQV | | Descriptor: | Beta-2-microglobulin, LEU-LEU-GLY-TRP-VAL-PHE-ALA-GLN-VAL, MHC class I antigen | | Authors: | Rizkallah, P.J, Man, S, Redman, J.E. | | Deposit date: | 2020-06-04 | | Release date: | 2021-06-30 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Synthetic Peptides with Inadvertent Chemical Modifications Can Activate Potentially Autoreactive T Cells.
J Immunol., 207, 2021
|
|
8V6V
 
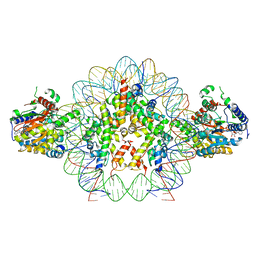 | | Cryo-EM structure of doubly-bound SNF2h-nucleosome complex | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Histone H2A type 1, Histone H2B, ... | | Authors: | Chio, U.S, Palovcak, E, Armache, J.P, Narlikar, G.J, Cheng, Y. | | Deposit date: | 2023-12-03 | | Release date: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | Functionalized graphene-oxide grids enable high-resolution cryo-EM structures of the SNF2h-nucleosome complex without crosslinking.
Nat Commun, 15, 2024
|
|
4YXW
 
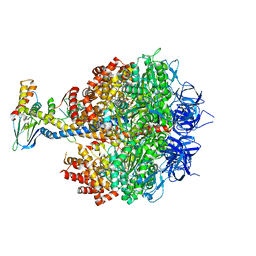 | | Bovine heart mitochondrial F1-ATPase inhibited by AMP-PNP and ADP in the presence of thiophosphate. | | Descriptor: | ATP synthase subunit alpha, mitochondrial, ATP synthase subunit beta, ... | | Authors: | Bason, J.V, Montgomery, M.G, Leslie, A.G.W, Walker, J.E. | | Deposit date: | 2015-03-23 | | Release date: | 2015-05-06 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | How release of phosphate from mammalian F1-ATPase generates a rotary substep.
Proc.Natl.Acad.Sci.USA, 112, 2015
|
|
