6HMT
 
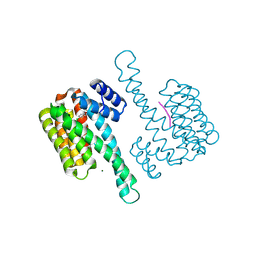 | | Ternary complex of Estrogen Receptor alpha peptide and 14-3-3 sigma C42 mutant bound to disulfide fragment PPI stabilizer 2 | | Descriptor: | 14-3-3 protein sigma, 2-(4-chloranylphenoxy)-2-methyl-~{N}-(2-sulfanylethyl)propanamide, Estrogen Receptor, ... | | Authors: | Sijbesma, E, Hallenbeck, K.K, Leysen, S, Arkin, M.R, Ottmann, C. | | Deposit date: | 2018-09-12 | | Release date: | 2019-02-27 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | Site-Directed Fragment-Based Screening for the Discovery of Protein-Protein Interaction Stabilizers.
J. Am. Chem. Soc., 141, 2019
|
|
1JEM
 
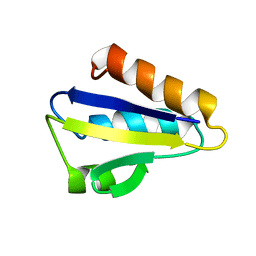 | | NMR STRUCTURE OF HISTIDINE PHOSPHORYLATED FORM OF THE PHOSPHOCARRIER HISTIDINE CONTAINING PROTEIN FROM BACILLUS SUBTILIS, NMR, 25 STRUCTURES | | Descriptor: | HISTIDINE CONTAINING PROTEIN | | Authors: | Jones, B.E, Rajagopal, P, Klevit, R.E. | | Deposit date: | 1997-04-01 | | Release date: | 1997-07-23 | | Last modified: | 2021-11-03 | | Method: | SOLUTION NMR | | Cite: | Phosphorylation on histidine is accompanied by localized structural changes in the phosphocarrier protein, HPr from Bacillus subtilis.
Protein Sci., 6, 1997
|
|
6TN5
 
 | | Rapid optimisation of fragments and hits to lead compounds from screening of crude reaction mixtures | | Descriptor: | CHLORIDE ION, Heat shock protein HSP 90-alpha, ~{N}-(4-aminocarbonylphenyl)-~{N}-methyl-2,4-bis(oxidanyl)benzamide | | Authors: | Baker, L.M, Aimon, A, Murray, J.B, Surgenor, A.E, Matassova, N, Roughley, S.D, von Delft, F, Hubbard, R.E. | | Deposit date: | 2019-12-05 | | Release date: | 2020-10-14 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (1.165 Å) | | Cite: | Rapid optimisation of fragments and hits to lead compounds from screening of crude reaction mixtures
Commun Chem, 2020
|
|
4QLU
 
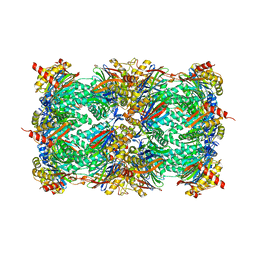 | | yCP in complex with tripeptidic epoxyketone inhibitor 9 | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, MAGNESIUM ION, N-[(3-methyl-1H-inden-2-yl)carbonyl]-D-alanyl-N-[(2S,4R)-1-cyclohexyl-5-hydroxy-4-methyl-3-oxopentan-2-yl]-L-tryptophanamide, ... | | Authors: | de Bruin, G, Huber, E, Xin, B, van Rooden, E, Al-Ayed, K, Kim, K, Kisselev, A, Driessen, C, van der Marel, G, Groll, M, Overkleeft, H. | | Deposit date: | 2014-06-13 | | Release date: | 2014-07-23 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structure-based design of beta 1i or beta 5i specific inhibitors of human immunoproteasomes
J.Med.Chem., 57, 2014
|
|
6HOL
 
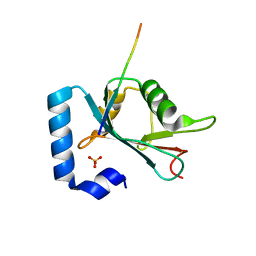 | | Structure of ATG14 LIR motif bound to GABARAPL1 | | Descriptor: | Beclin 1-associated autophagy-related key regulator, GLYCEROL, Gamma-aminobutyric acid receptor-associated protein-like 1, ... | | Authors: | Mouilleron, S, Birgisdottir, A.B, Bhujbal, Z, Wirth, M, Sjottem, E, Evjen, G, Zhang, W, Lee, R, O'Reilly, N, Tooze, S, Lamark, T, Johansen, T. | | Deposit date: | 2018-09-17 | | Release date: | 2019-02-27 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Members of the autophagy class III phosphatidylinositol 3-kinase complex I interact with GABARAP and GABARAPL1 via LIR motifs.
Autophagy, 15, 2019
|
|
6REG
 
 | | Crystal structure of Pizza6-S with Zn2+ | | Descriptor: | GLYCEROL, Pizza6-S, ZINC ION | | Authors: | Noguchi, H, Clarke, D.E, Gryspeerdt, J.L, Feyter, S.D, Voet, A.R.D. | | Deposit date: | 2019-04-12 | | Release date: | 2019-07-31 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Artificial beta-propeller protein-based hydrolases.
Chem.Commun.(Camb.), 55, 2019
|
|
6REK
 
 | | Crystal structure of Pizza6-SH with Cu2+ | | Descriptor: | COPPER (II) ION, GLYCEROL, Pizza6-SH | | Authors: | Noguchi, H, Clarke, D.E, Gryspeerdt, J.L, Feyter, S.D, Voet, A.R.D. | | Deposit date: | 2019-04-12 | | Release date: | 2019-07-31 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | Artificial beta-propeller protein-based hydrolases.
Chem.Commun.(Camb.), 55, 2019
|
|
1U7T
 
 | | Crystal Structure of ABAD/HSD10 with a Bound Inhibitor | | Descriptor: | 1-AZEPAN-1-YL-2-PHENYL-2-(4-THIOXO-1,4-DIHYDRO-PYRAZOLO[3,4-D]PYRIMIDIN-5-YL)ETHANONE ADDUCT, 3-hydroxyacyl-CoA dehydrogenase type II, NICOTINAMIDE-ADENINE-DINUCLEOTIDE | | Authors: | Kissinger, C.R, Rejto, P.A, Pelletier, L.A, Showalter, R.E, Villafranca, J.E. | | Deposit date: | 2004-08-04 | | Release date: | 2004-10-05 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of human ABAD/HSD10 with a bound inhibitor: implications for design of Alzheimer's disease therapeutics
J.Mol.Biol., 342, 2004
|
|
6Z0Q
 
 | | Crystal structure of SAICAR Synthetase (PurC) from Mycobacterium abscessus in complex with fragment 2 | | Descriptor: | Phosphoribosylaminoimidazole-succinocarboxamide synthase, SULFATE ION, ethyl (2~{S},3~{R})-3-(5-bromanylpyridin-2-yl)-2-fluoranyl-3-oxidanyl-propanoate | | Authors: | Thomas, S.E, Charoensutthivarakul, S, Coyne, A.G, Abell, C, Blundell, T.L. | | Deposit date: | 2020-05-10 | | Release date: | 2021-05-19 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.535 Å) | | Cite: | Development of Inhibitors of SAICAR Synthetase (PurC) from Mycobacterium abscessus Using a Fragment-Based Approach.
Acs Infect Dis., 8, 2022
|
|
6HQ8
 
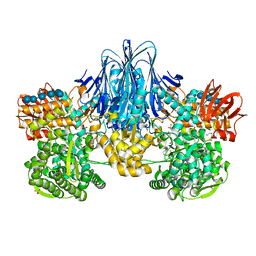 | | Bacterial beta-1,3-oligosaccharide phosphorylase from GH149 with laminarihexaose bound at a surface site | | Descriptor: | 1,2-ETHANEDIOL, BICINE, Beta-1,3-oligosaccharide phosphorylase, ... | | Authors: | Kuhaudomlarp, S, Stevenson, C.E.M, Lawson, D.M, Field, R.A. | | Deposit date: | 2018-09-24 | | Release date: | 2019-06-12 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | The structure of a GH149 beta-(1 → 3) glucan phosphorylase reveals a new surface oligosaccharide binding site and additional domains that are absent in the disaccharide-specific GH94 glucose-beta-(1 → 3)-glucose (laminaribiose) phosphorylase.
Proteins, 87, 2019
|
|
7TL3
 
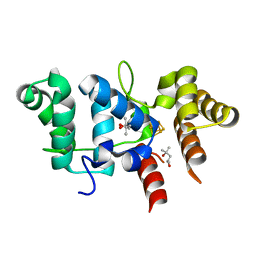 | | Crystal Structure of Yeast p58C Multi-Tyrosine Mutant 5YF431 | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, DNA primase large subunit, IRON/SULFUR CLUSTER | | Authors: | Blee, A.M, Salay, L.E, Chazin, W.J. | | Deposit date: | 2022-01-18 | | Release date: | 2022-06-29 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.066 Å) | | Cite: | Modification of the 4Fe-4S Cluster Charge Transport Pathway Alters RNA Synthesis by Yeast DNA Primase.
Biochemistry, 61, 2022
|
|
7U0K
 
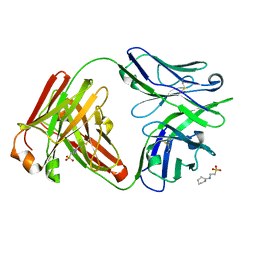 | | IOMA class antibody Fab ACS124 | | Descriptor: | 2-[N-CYCLOHEXYLAMINO]ETHANE SULFONIC ACID, IOMA Class antibody ACS124 Heavy chain, IOMA Class antibody ACS124 Light chain | | Authors: | Farokhi, E, Stanfield, R.L, Wilson, I.A. | | Deposit date: | 2022-02-18 | | Release date: | 2022-08-24 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.73 Å) | | Cite: | Identification of IOMA-class neutralizing antibodies targeting the CD4-binding site on the HIV-1 envelope glycoprotein.
Nat Commun, 13, 2022
|
|
7XA3
 
 | | Cryo-EM structure of the CCL2 bound CCR2-Gi complex | | Descriptor: | C-C motif chemokine 2, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | Authors: | Shao, Z, Tan, Y, Shen, Q, Yao, B, Hou, L, Qin, J, Xu, P, Mao, C, Chen, L, Zhang, H, Shen, D, Zhang, C, Li, W, Du, X, Li, F, Chen, Z, Jiang, Y, Xu, H.E, Ying, S, Ma, H, Zhang, Y, Shen, H. | | Deposit date: | 2022-03-17 | | Release date: | 2022-08-24 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Molecular insights into ligand recognition and activation of chemokine receptors CCR2 and CCR3.
Cell Discov, 8, 2022
|
|
6HRG
 
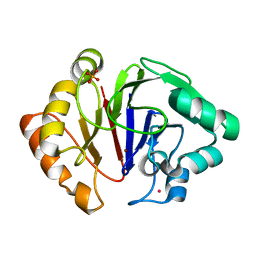 | | Structure of Igni18, a novel metallo hydrolase from the hyperthermophilic archaeon Ignicoccus hospitalis KIN4/I | | Descriptor: | PHOSPHATE ION, POTASSIUM ION, UPF0173 metal-dependent hydrolase Igni_1254, ... | | Authors: | Smits, S.H, Streit, W.R, Jaeger, K.E, Hoeppner, A. | | Deposit date: | 2018-09-26 | | Release date: | 2019-10-09 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (2.12 Å) | | Cite: | A promiscuous ancestral enzyme ́s structure unveils protein variable regions of the highly diverse metallo-beta-lactamase family.
Commun Biol, 4, 2021
|
|
2J9I
 
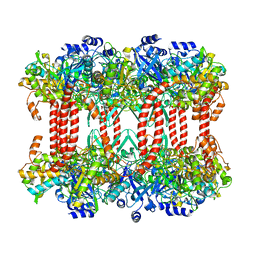 | | Lengsin is a survivor of an ancient family of class I glutamine synthetases in eukaryotes that has undergone evolutionary re- engineering for a tissue-specific role in the vertebrate eye lens. | | Descriptor: | GLUTAMATE-AMMONIA LIGASE DOMAIN-CONTAINING PROTEIN 1 | | Authors: | Wyatt, K, White, H.E, Wang, L, Bateman, O.A, Slingsby, C, Orlova, E.V, Wistow, G. | | Deposit date: | 2006-11-09 | | Release date: | 2006-12-13 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (17 Å) | | Cite: | Lengsin is a Survivor of an Ancient Family of Class I Glutamine Synthetases Re-Engineered by Evolution for a Role in the Vertebrate Lens.
Structure, 14, 2006
|
|
7X9Y
 
 | | Cryo-EM structure of the apo CCR3-Gi complex | | Descriptor: | C-C chemokine receptor type 3, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | Authors: | Shao, Z, Tan, Y, Shen, Q, Yao, B, Hou, L, Qin, J, Xu, P, Mao, C, Chen, L, Zhang, H, Shen, D, Zhang, C, Li, W, Du, X, Li, F, Chen, Z, Jiang, Y, Xu, H.E, Ying, S, Ma, H, Zhang, Y, Shen, H. | | Deposit date: | 2022-03-16 | | Release date: | 2022-08-24 | | Last modified: | 2024-06-26 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Molecular insights into ligand recognition and activation of chemokine receptors CCR2 and CCR3.
Cell Discov, 8, 2022
|
|
7TTI
 
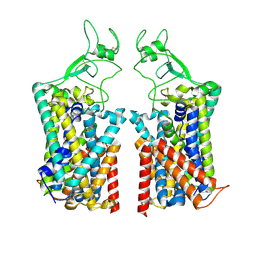 | | Human KCC1 bound with VU0463271 In an outward-open state | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, N-cyclopropyl-N-(4-methyl-1,3-thiazol-2-yl)-2-[(6-phenylpyridazin-3-yl)sulfanyl]acetamide, Solute carrier family 12 member 4 | | Authors: | Zhao, Y.X, Cao, E.H. | | Deposit date: | 2022-02-01 | | Release date: | 2022-06-29 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Structure of the human cation-chloride cotransport KCC1 in an outward-open state.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
6E0L
 
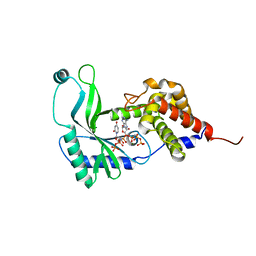 | | Structure of Rhodothermus marinus CdnE c-UMP-AMP synthase with Apcpp and Upnpp | | Descriptor: | 5'-O-[(S)-hydroxy{[(S)-hydroxy(phosphonooxy)phosphoryl]amino}phosphoryl]uridine, DIPHOSPHOMETHYLPHOSPHONIC ACID ADENOSYL ESTER, MAGNESIUM ION, ... | | Authors: | Eaglesham, J.B, Whiteley, A.T, de Oliveira Mann, C.C, Morehouse, B.R, Nieminen, E.A, King, D.S, Lee, A.S.Y, Mekalanos, J.J, Kranzusch, P.J. | | Deposit date: | 2018-07-06 | | Release date: | 2019-02-20 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Bacterial cGAS-like enzymes synthesize diverse nucleotide signals.
Nature, 567, 2019
|
|
6YYB
 
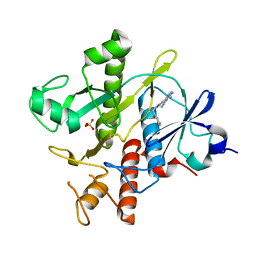 | | Crystal structure of SAICAR Synthetase (PurC) from Mycobacterium abscessus in complex with inhibitor | | Descriptor: | 4-azanyl-6-[1-[(1~{R})-1-phenylethyl]pyrazol-4-yl]pyrimidine-5-carbonitrile, Phosphoribosylaminoimidazole-succinocarboxamide synthase, SULFATE ION | | Authors: | Thomas, S.E, Charoensutthivarakul, S, Coyne, A.G, Abell, C, Blundell, T.L. | | Deposit date: | 2020-05-04 | | Release date: | 2021-05-12 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.51 Å) | | Cite: | Development of Inhibitors of SAICAR Synthetase (PurC) from Mycobacterium abscessus Using a Fragment-Based Approach.
Acs Infect Dis., 8, 2022
|
|
6ZHG
 
 | | Ca2+-ATPase from Listeria Monocytogenes in complex with AlF | | Descriptor: | Calcium-transporting ATPase, MAGNESIUM ION, TETRAFLUOROALUMINATE ION | | Authors: | Basse Hansen, S, Dyla, M, Neumann, C, Quistgaard, E.M.H, Lauwring Andersen, J, Kjaergaard, M, Nissen, P. | | Deposit date: | 2020-06-23 | | Release date: | 2021-05-19 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (4 Å) | | Cite: | The Crystal Structure of the Ca 2+ -ATPase 1 from Listeria monocytogenes reveals a Pump Primed for Dephosphorylation.
J.Mol.Biol., 433, 2021
|
|
6Z0Z
 
 | | Human wtSTING in complex with 3',3'-c-(2'FdAMP-2'FdAMP) | | Descriptor: | 2'-fluoro-,3',3'-c-di-AMP, Stimulator of interferon protein | | Authors: | Boura, E, Smola, M. | | Deposit date: | 2020-05-11 | | Release date: | 2021-05-19 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.499 Å) | | Cite: | Ligand Strain and Its Conformational Complexity Is a Major Factor in the Binding of Cyclic Dinucleotides to STING Protein.
Angew.Chem.Int.Ed.Engl., 60, 2021
|
|
6TT3
 
 | |
6R8S
 
 | | Crystal structure of aIF2gamma subunit I181K from archaeon Sulfolobus solfataricus complexed with GDPCP | | Descriptor: | FORMIC ACID, MAGNESIUM ION, PHOSPHOMETHYLPHOSPHONIC ACID GUANYLATE ESTER, ... | | Authors: | Kravchenko, O, Arkhipova, V, Gabdulkhakov, A, Stolboushkina, E, Nikonov, O, Garber, M, Nikonov, S. | | Deposit date: | 2019-04-02 | | Release date: | 2020-05-06 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.18 Å) | | Cite: | Crystal structure of aIF2gamma subunit I181K from archaeon Sulfolobus solfataricus complexed with GDPCP
To Be Published
|
|
7TL2
 
 | | Crystal Structure of Yeast p58C Multi-Tyrosine Mutant 5YF412 | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, DNA primase large subunit, GLYCEROL, ... | | Authors: | Blee, A.M, Salay, L.E, Chazin, W.J. | | Deposit date: | 2022-01-18 | | Release date: | 2022-06-29 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.529 Å) | | Cite: | Modification of the 4Fe-4S Cluster Charge Transport Pathway Alters RNA Synthesis by Yeast DNA Primase.
Biochemistry, 61, 2022
|
|
6DZW
 
 | | Cryo-EM structure of the ts2-inactive human serotonin transporter in complex with paroxetine and 15B8 Fab and 8B6 ScFv | | Descriptor: | 15B8 antibody heavy chain, 15B8 antibody light chain, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Coleman, J.A, Yang, D, Gouaux, E. | | Deposit date: | 2018-07-05 | | Release date: | 2019-04-24 | | Last modified: | 2024-10-09 | | Method: | ELECTRON MICROSCOPY (4.3 Å) | | Cite: | Serotonin transporter-ibogaine complexes illuminate mechanisms of inhibition and transport.
Nature, 569, 2019
|
|
