6YJ2
 
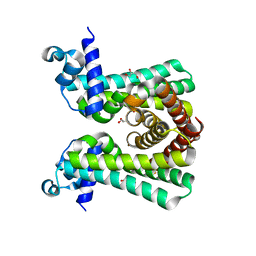 | | Structural and DNA binding studies of the transcriptional repressor Rv2506 (BkaR) from Mycobacterium tuberculosis supports a role in L-Leucine catabolism | | Descriptor: | GLYCEROL, Probable transcriptional regulatory protein (Probably TetR-family) | | Authors: | Keep, N.H, Pritchard, J.E, Sula, A, Cole, A.R, Kendall, S.L. | | Deposit date: | 2020-04-02 | | Release date: | 2021-04-14 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural and DNA binding studies of the transcriptional repressor Rv2506 (BkaR) from Mycobacterium tuberculosis supports a role in L-Leucine catabolism
To be published
|
|
7SJN
 
 | | HtrA1:Fab15H6.v4 complex | | Descriptor: | Fab15H6.v4 Heavy Chain, Fab15H6.v4 Light Chain, Serine protease HTRA1 | | Authors: | Gerhardy, S, Green, E, Estevez, A, Arthur, C.P, Ultsch, M, Rohou, A, Kirchhofer, D. | | Deposit date: | 2021-10-18 | | Release date: | 2022-09-07 | | Last modified: | 2022-09-21 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Allosteric inhibition of HTRA1 activity by a conformational lock mechanism to treat age-related macular degeneration.
Nat Commun, 13, 2022
|
|
7RWY
 
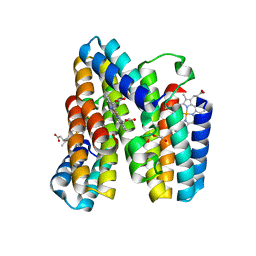 | |
4ZCS
 
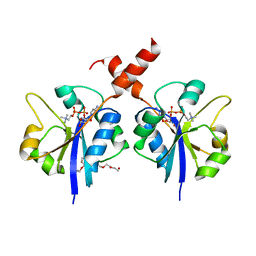 | | Crystal structure of the C-terminal catalytic domain of Plasmodium falciparum CTP:phosphocholine cytidylyltransferase in complex with CDP-choline | | Descriptor: | 3,6,9,12,15,18-HEXAOXAICOSANE-1,20-DIOL, Choline-phosphate cytidylyltransferase, [2-CYTIDYLATE-O'-PHOSPHONYLOXYL]-ETHYL-TRIMETHYL-AMMONIUM | | Authors: | Guca, E, Hoh, F, Guichou, J.-F, Cerdan, R. | | Deposit date: | 2015-04-16 | | Release date: | 2016-08-17 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Structural determinants of the catalytic mechanism of Plasmodium CCT, a key enzyme of malaria lipid biosynthesis.
Sci Rep, 8, 2018
|
|
2JU4
 
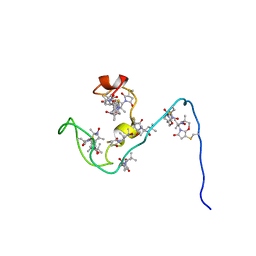 | | NMR structure of the gamma subunit of cGMP phosphodiesterase | | Descriptor: | (3'R)-1'-oxyl-2',2',5',5'-tetramethyl-1,3'-bipyrrolidine-2,5-dione, Retinal rod rhodopsin-sensitive cGMP 3',5'-cyclic phosphodiesterase subunit gamma | | Authors: | Song, J, Guo, L.W, Ruoho, A.E, Markley, J.L, Center for Eukaryotic Structural Genomics (CESG) | | Deposit date: | 2007-08-14 | | Release date: | 2007-10-16 | | Last modified: | 2020-02-19 | | Method: | SOLUTION NMR | | Cite: | Intrinsically disordered gamma-subunit of cGMP phosphodiesterase encodes functionally relevant transient secondary and tertiary structure.
Proc.Natl.Acad.Sci.USA, 105, 2008
|
|
7SKT
 
 | |
2C3Y
 
 | | CRYSTAL STRUCTURE OF THE RADICAL FORM OF PYRUVATE:FERREDOXIN OXIDOREDUCTASE FROM Desulfovibrio africanus | | Descriptor: | 2-ACETYL-THIAMINE DIPHOSPHATE, CALCIUM ION, CARBON DIOXIDE, ... | | Authors: | Cavazza, C, Contreras-Martel, C, Pieulle, L, Chabriere, E, Hatchikian, E.C, Fontecilla-Camps, J.C. | | Deposit date: | 2005-10-13 | | Release date: | 2006-02-15 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.93 Å) | | Cite: | Flexibility of Thiamine Diphosphate Revealed by Kinetic Crystallographic Studies of the Reaction of Pyruvate-Ferredoxin Oxidoreductase with Pyruvate.
Structure, 14, 2006
|
|
6W67
 
 | |
6YDD
 
 | | X-ray structure of LPMO. | | Descriptor: | COPPER (II) ION, LPMO lytic polysaccharide monooxygenase, beta-D-glucopyranose-(1-4)-beta-D-glucopyranose, ... | | Authors: | Tandrup, T, Tryfona, T, Frandsen, K.E.H, Johansen, K.S, Dupree, P, Lo Leggio, L. | | Deposit date: | 2020-03-20 | | Release date: | 2020-09-16 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Oligosaccharide Binding and Thermostability of Two Related AA9 Lytic Polysaccharide Monooxygenases.
Biochemistry, 59, 2020
|
|
4GC9
 
 | | Crystal structure of murine TFB1M in complex with SAM | | Descriptor: | ACETATE ION, Dimethyladenosine transferase 1, mitochondrial, ... | | Authors: | Guja, K.E, Yakubovskaya, E, Shi, H, Mejia, E, Hambardjieva, E, Venkataraman, K, Karzai, A.W, Garcia-Diaz, M. | | Deposit date: | 2012-07-30 | | Release date: | 2013-07-10 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (2.103 Å) | | Cite: | Structural basis for S-adenosylmethionine binding and methyltransferase activity by mitochondrial transcription factor B1.
Nucleic Acids Res., 41, 2013
|
|
1RQ7
 
 | | MYCOBACTERIUM TUBERCULOSIS FTSZ IN COMPLEX WITH GDP | | Descriptor: | Cell division protein ftsZ, GUANOSINE-5'-DIPHOSPHATE | | Authors: | Leung, A.K.W, White, E.L, Ross, L.J, Reynolds, R.C, DeVito, J.A, Borhani, D.W. | | Deposit date: | 2003-12-04 | | Release date: | 2004-08-31 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structure of Mycobacterium tuberculosis FtsZ reveals unexpected, G protein-like conformational switches.
J.Mol.Biol., 342, 2004
|
|
4PLE
 
 | | Human Nuclear Receptor Liver Receptor Homologue-1, LRH-1, Bound to an E. Coli Phospholipid and a Fragment of TIF-2 | | Descriptor: | 3-[(3-CHOLAMIDOPROPYL)DIMETHYLAMMONIO]-1-PROPANESULFONATE, L-ALPHA-PHOSPHATIDYL-BETA-OLEOYL-GAMMA-PALMITOYL-PHOSPHATIDYLETHANOLAMINE, Nuclear receptor coactivator 2, ... | | Authors: | Ortlund, E.A, Musille, P.M. | | Deposit date: | 2014-05-16 | | Release date: | 2015-12-16 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.752 Å) | | Cite: | Unexpected Allosteric Network Contributes to LRH-1 Co-regulator Selectivity.
J.Biol.Chem., 291, 2016
|
|
5E4A
 
 | | Human transthyretin (TTR) complexed with (2,7-Dichloro-fluoren-9-ylideneaminooxy)-acetic acid. | | Descriptor: | Transthyretin, {[(2,7-dichloro-9H-fluoren-9-ylidene)amino]oxy}acetic acid | | Authors: | Ciccone, L, Nencetti, S, Rossello, A, Orlandini, E, Stura, E.A. | | Deposit date: | 2015-10-05 | | Release date: | 2016-03-23 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.33 Å) | | Cite: | Synthesis and structural analysis of halogen substituted fibril formation inhibitors of Human Transthyretin (TTR).
J Enzyme Inhib Med Chem, 31, 2016
|
|
1HQF
 
 | | CRYSTAL STRUCTURE OF THE BINUCLEAR MANGANESE METALLOENZYME ARGINASE COMPLEXED WITH N-HYDROXY-L-ARGININE | | Descriptor: | ARGINASE 1, MANGANESE (II) ION, N-OMEGA-HYDROXY-L-ARGININE | | Authors: | Cox, J.D, Cama, E, Colleluori, D.M, Ash, D.E, Christianson, D.W. | | Deposit date: | 2000-12-16 | | Release date: | 2001-04-04 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Mechanistic and metabolic inferences from the binding of substrate analogues and products to arginase.
Biochemistry, 40, 2001
|
|
5E6A
 
 | | Glucocorticoid receptor DNA binding domain - PLAU NF-kB response element complex | | Descriptor: | DNA (5'-D(*AP*TP*CP*AP*GP*GP*AP*AP*AP*TP*TP*CP*CP*CP*AP*G)-3'), DNA (5'-D(*CP*TP*GP*GP*GP*AP*AP*TP*TP*TP*CP*CP*TP*GP*AP*T)-3'), Glucocorticoid receptor, ... | | Authors: | Hudson, W.H, Rye, E.A, Herbst, A.G, Ortlund, E.A. | | Deposit date: | 2015-10-09 | | Release date: | 2017-02-08 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.197 Å) | | Cite: | Cryptic glucocorticoid receptor-binding sites pervade genomic NF-kappa B response elements.
Nat Commun, 9, 2018
|
|
6W66
 
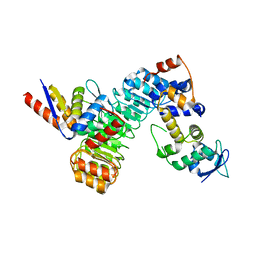 | | The structure of the F64A, S172A mutant Keap1-BTB domain in complex with SKP1-FBXL17 | | Descriptor: | F-box/LRR-repeat protein 17, Kelch-like ECH-associated protein 1, S-phase kinase-associated protein 1 | | Authors: | Mena, E.L, Gee, C.L, Kuriyan, J, Rape, M. | | Deposit date: | 2020-03-16 | | Release date: | 2020-08-19 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (3.21 Å) | | Cite: | Structural basis for dimerization quality control.
Nature, 586, 2020
|
|
6YOE
 
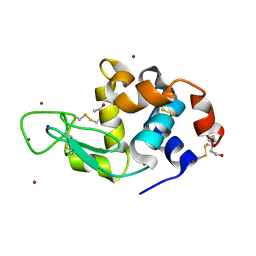 | | Structure of Lysozyme from SiN IMISX setup collected by still serial crystallography on crystals prelocated by 2D X-ray phase-contrast imaging | | Descriptor: | 2-(2-ETHOXYETHOXY)ETHANOL, ACETIC ACID, BROMIDE ION, ... | | Authors: | Huang, C.-Y, Martiel, I, Villanueva-Perez, P, Panepucci, E, Caffrey, M, Wang, M. | | Deposit date: | 2020-04-14 | | Release date: | 2020-11-04 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Low-dose in situ prelocation of protein microcrystals by 2D X-ray phase-contrast imaging for serial crystallography.
Iucrj, 7, 2020
|
|
5YT1
 
 | |
5EA0
 
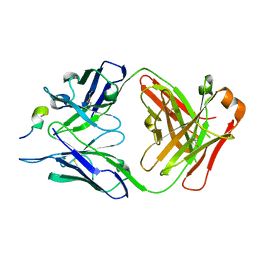 | | Structure of the antibody 7968 with human complement factor H-derived peptide | | Descriptor: | Complement factor H-related protein 2, Heavy chain of antibody 7968 Fab fragment, Light chain of antibody 7968 Fab fragment | | Authors: | Bushey, R.T, Moody, M.A, Nicely, N.I, Alam, S.M, Haynes, B.F, Winkler, M.T, Gottlin, E.B, Campa, M.J, Liao, H.-X, Patz Jr, E.F. | | Deposit date: | 2015-10-15 | | Release date: | 2016-05-25 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | A Therapeutic Antibody for Cancer, Derived from Single Human B Cells.
Cell Rep, 15, 2016
|
|
6BR7
 
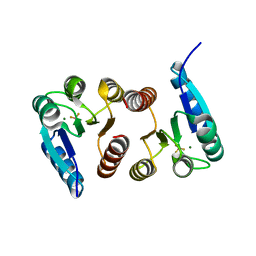 | |
226L
 
 | | GENERATING LIGAND BINDING SITES IN T4 LYSOZYME USING DEFICIENCY-CREATING SUBSTITUTIONS | | Descriptor: | BETA-MERCAPTOETHANOL, CHLORIDE ION, T4 LYSOZYME | | Authors: | Baldwin, E.P, Baase, W.A, Zhang, X.-J, Feher, V, Matthews, B.W. | | Deposit date: | 1997-06-25 | | Release date: | 1998-03-18 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Generation of ligand binding sites in T4 lysozyme by deficiency-creating substitutions.
J.Mol.Biol., 277, 1998
|
|
6Y8N
 
 | | Mycobacterium thermoresistibile GyrB21 in complex with Redx03863 | | Descriptor: | 1,2-ETHANEDIOL, 4-[(1~{S},5~{R})-6-azanyl-3-azabicyclo[3.1.0]hexan-3-yl]-6-fluoranyl-~{N}-methyl-2-(2-methylpyrimidin-5-yl)oxy-9~{H}-pyrimido[4,5-b]indol-8-amine, DNA gyrase subunit B, ... | | Authors: | Henderson, S.R, Stevenson, C.E.M, Malone, B, Zholnerovych, Y, Mitchenall, L.A, Pichowicz, M, McGarry, D.H, Cooper, I.R, Charrier, C, Salisbury, A, Lawson, D.M, Maxwell, A. | | Deposit date: | 2020-03-05 | | Release date: | 2020-08-12 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structural and mechanistic analysis of ATPase inhibitors targeting mycobacterial DNA gyrase.
J.Antimicrob.Chemother., 75, 2020
|
|
4Z0V
 
 | | The structure of human PDE12 residues 161-609 | | Descriptor: | 2',5'-phosphodiesterase 12, GLYCEROL, MAGNESIUM ION | | Authors: | Nolte, R.T, Wisely, B, Wang, L, Wood, E.R. | | Deposit date: | 2015-03-26 | | Release date: | 2015-06-17 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.78 Å) | | Cite: | The Role of Phosphodiesterase 12 (PDE12) as a Negative Regulator of the Innate Immune Response and the Discovery of Antiviral Inhibitors.
J.Biol.Chem., 290, 2015
|
|
6Y4J
 
 | | Engineered Fructosyl Peptide Oxidase | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, Fructosyl Peptide Oxidase, GLYCEROL, ... | | Authors: | Donini, S, Rigoldi, F, Gautieri, A, Parisini, E. | | Deposit date: | 2020-02-21 | | Release date: | 2021-01-13 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.38 Å) | | Cite: | Rational backbone redesign of a fructosyl peptide oxidase to widen its active site access tunnel.
Biotechnol.Bioeng., 117, 2020
|
|
6VTK
 
 | |
