7QEE
 
 | | SN243 mutant D415N bound to para-nitrophenyl-Beta-D-glucuronide | | Descriptor: | 4-nitrophenyl beta-D-glucopyranosiduronic acid, SN243, SULFATE ION, ... | | Authors: | Neun, S, Brear, P, Campbell, E, Omari, K, Wagner, O, Hyvonen, M, Hollfelder, F. | | Deposit date: | 2021-12-02 | | Release date: | 2022-11-16 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.374 Å) | | Cite: | Functional metagenomic screening identifies an unexpected beta-glucuronidase.
Nat.Chem.Biol., 18, 2022
|
|
7H9B
 
 | | Group deposition for crystallographic fragment screening of Chikungunya virus nsP3 macrodomain -- Crystal structure of Chikungunya virus nsP3 macrodomain in complex with Z1201619441 (CHIKV_MacB-x1417) | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 3,5-DIMETHYL-1H-PYRAZOLE-4-CARBOXYLIC ACID ETHYL ESTER, CHLORIDE ION, ... | | Authors: | Aschenbrenner, J.C, Fairhead, M, Godoy, A.S, Balcomb, B.H, Capkin, E, Chandran, A.V, Dolci, I, Golding, M, Koekemoer, L, Lithgo, R.M, Marples, P.G, Ni, X, Oliva, G, Thompson, W, Tomlinson, C.W.E, Wild, C, Winokan, M, Xavier, M.-A.E, Fearon, D, von Delft, F. | | Deposit date: | 2024-04-26 | | Release date: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (1.61 Å) | | Cite: | Group deposition for crystallographic fragment screening of Chikungunya virus nsP3 macrodomain
To Be Published
|
|
5IC6
 
 | | Crystal structure of caspase-7 DEVE peptide complex | | Descriptor: | Caspase-7 subunit p11, Caspase-7 subunit p20, DEVE peptide | | Authors: | Seaman, J.E, Julien, O, Lee, P.S, Rettenmaier, T.J, Thomsen, N.D, Wells, J.A. | | Deposit date: | 2016-02-22 | | Release date: | 2016-07-20 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Cacidases: caspases can cleave after aspartate, glutamate and phosphoserine residues.
Cell Death Differ., 23, 2016
|
|
8T3I
 
 | | Crystal structure of mutant exfoliative toxin C (ExhC) from Mammaliicoccus sciuri | | Descriptor: | Exfoliative toxin C | | Authors: | Gismene, C, Calil, F.A, Hernandez Gonzalez, J.E, Ziem Nascimento, A.F, Santisteban, A.R.N, Arni, R.K, Barros Mariutti, R. | | Deposit date: | 2023-06-07 | | Release date: | 2023-11-08 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.57 Å) | | Cite: | Necrotic activity of ExhC from Mammaliicoccus sciuri is mediated by specific amino acid residues.
Int.J.Biol.Macromol., 254, 2024
|
|
7H4J
 
 | | Group deposition for crystallographic fragment screening of Coxsackievirus A16 (G-10) 2A protease -- Crystal structure of Coxsackievirus A16 (G-10) 2A protease in complex with Z53825020 (A71EV2A-x0875) | | Descriptor: | 1-(ethanesulfonyl)piperidine-4-carboxylic acid, DIMETHYL SULFOXIDE, Protease 2A, ... | | Authors: | Lithgo, R.M, Fairhead, M, Koekemoer, L, Balcomb, B.H, Capkin, E, Chandran, A.V, Golding, M, Godoy, A.S, Aschenbrenner, J.C, Marples, P.G, Ni, X, Thompson, W, Tomlinson, C.W.E, Wild, C, Winokan, M, Xavier, M.-A.E, Fearon, D, von Delft, F. | | Deposit date: | 2024-04-04 | | Release date: | 2024-04-24 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.79 Å) | | Cite: | Crystallographic Fragment Screen of Coxsackievirus A16 2A Protease identifies new opportunities for the development of broad-spectrum anti-enterovirals.
Biorxiv, 2024
|
|
7H4W
 
 | | Group deposition for crystallographic fragment screening of Coxsackievirus A16 (G-10) 2A protease -- Crystal structure of Coxsackievirus A16 (G-10) 2A protease in complex with Z56042334 (A71EV2A-x1140) | | Descriptor: | 1-(2-chlorophenyl)-2-(pyrrolidin-1-yl)ethan-1-one, DIMETHYL SULFOXIDE, Protease 2A, ... | | Authors: | Lithgo, R.M, Fairhead, M, Koekemoer, L, Balcomb, B.H, Capkin, E, Chandran, A.V, Golding, M, Godoy, A.S, Aschenbrenner, J.C, Marples, P.G, Ni, X, Thompson, W, Tomlinson, C.W.E, Wild, C, Winokan, M, Xavier, M.-A.E, Fearon, D, von Delft, F. | | Deposit date: | 2024-04-04 | | Release date: | 2024-04-24 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.52 Å) | | Cite: | Crystallographic Fragment Screen of Coxsackievirus A16 2A Protease identifies new opportunities for the development of broad-spectrum anti-enterovirals.
Biorxiv, 2024
|
|
2HMQ
 
 | |
7H4Y
 
 | | Group deposition for crystallographic fragment screening of Coxsackievirus A16 (G-10) 2A protease -- Crystal structure of Coxsackievirus A16 (G-10) 2A protease in complex with Z362043378 (A71EV2A-x1146) | | Descriptor: | 2-phenylpyrimidin-5-ol, DIMETHYL SULFOXIDE, Protease 2A, ... | | Authors: | Lithgo, R.M, Fairhead, M, Koekemoer, L, Balcomb, B.H, Capkin, E, Chandran, A.V, Golding, M, Godoy, A.S, Aschenbrenner, J.C, Marples, P.G, Ni, X, Thompson, W, Tomlinson, C.W.E, Wild, C, Winokan, M, Xavier, M.-A.E, Fearon, D, von Delft, F. | | Deposit date: | 2024-04-04 | | Release date: | 2024-04-24 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Crystallographic Fragment Screen of Coxsackievirus A16 2A Protease identifies new opportunities for the development of broad-spectrum anti-enterovirals.
Biorxiv, 2024
|
|
1TTP
 
 | | TRYPTOPHAN SYNTHASE (E.C.4.2.1.20) IN THE PRESENCE OF CESIUM, ROOM TEMPERATURE | | Descriptor: | CESIUM ION, PYRIDOXAL-5'-PHOSPHATE, TRYPTOPHAN SYNTHASE | | Authors: | Rhee, S, Parris, K, Ahmed, S, Miles, E.W, Davies, D.R. | | Deposit date: | 1995-10-11 | | Release date: | 1996-03-08 | | Last modified: | 2024-06-05 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Exchange of K+ or Cs+ for Na+ induces local and long-range changes in the three-dimensional structure of the tryptophan synthase alpha2beta2 complex.
Biochemistry, 35, 1996
|
|
8JGF
 
 | | CryoEM structure of Gq-coupled MRGPRX1 with peptide agonist BAM8-22 | | Descriptor: | BAM8-22, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | Authors: | Sun, J.P, Xu, H.E, Yang, F, Liu, Z.M, Guo, L.L, Zhang, Y.M, Fang, G.X, Tie, L, Zhuang, Y.M, Xue, C.Y. | | Deposit date: | 2023-05-20 | | Release date: | 2024-01-10 | | Last modified: | 2024-11-13 | | Method: | ELECTRON MICROSCOPY (2.7 Å) | | Cite: | Ligand recognition and G protein coupling of the human itch receptor MRGPRX1.
Nat Commun, 14, 2023
|
|
6T8X
 
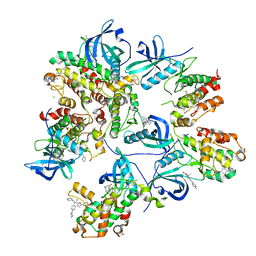 | | Crystal structure of MAPKAPK2 (MK2) complexed with PF-3644022 and 5-(4-bromophenyl)-N-[4-(1-piperazinyl)phenyl]-N-(2-pyridinylmethyl)-2-furancarboxamide | | Descriptor: | (10R)-10-methyl-3-(6-methylpyridin-3-yl)-9,10,11,12-tetrahydro-8H-[1,4]diazepino[5',6':4,5]thieno[3,2-f]quinolin-8-one, 5-(4-bromophenyl)-~{N}-(4-piperazin-1-ylphenyl)-~{N}-(pyridin-2-ylmethyl)furan-2-carboxamide, CHLORIDE ION, ... | | Authors: | Beaumont, E.J, Barker, J. | | Deposit date: | 2019-10-25 | | Release date: | 2021-05-12 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.81 Å) | | Cite: | Crystal structure of MAPKAPK2 (MK2) complexed with PF-3644022 and 5-(4-bromophenyl)-N-[4-(1-piperazinyl)phenyl]-N-(2-pyridinylmethyl)-2-furancarboxamide
To Be Published
|
|
6KZA
 
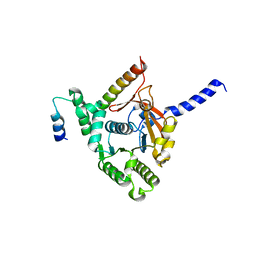 | | Crystal structure of the complex of the interaction domains of E. coli DnaB helicase and DnaC helicase loader | | Descriptor: | DNA replication protein DnaC, Replicative DNA helicase | | Authors: | Nagata, K, Okada, A, Ohtsuka, J, Ohkuri, T, Akama, Y, Sakiyama, Y, Miyazaki, E, Horita, S, Katayama, T, Ueda, T, Tanokura, M. | | Deposit date: | 2019-09-23 | | Release date: | 2019-11-20 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Crystal structure of the complex of the interaction domains of Escherichia coli DnaB helicase and DnaC helicase loader: structural basis implying a distortion-accumulation mechanism for the DnaB ring opening caused by DnaC binding.
J.Biochem., 167, 2020
|
|
8A2C
 
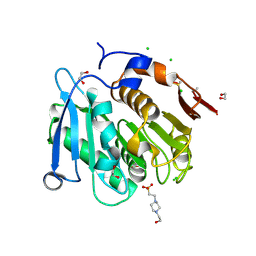 | | The crystal structure of the S178A mutant of PET40, a PETase enzyme from an unclassified Amycolatopsis | | Descriptor: | 1,2-ETHANEDIOL, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, CHLORIDE ION, ... | | Authors: | Costanzi, E, Applegate, V, Port, A, Smits, S.H.J. | | Deposit date: | 2022-06-03 | | Release date: | 2023-06-14 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | The metagenome-derived esterase PET40 is highly promiscuous and hydrolyses polyethylene terephthalate (PET).
Febs J., 291, 2024
|
|
2HMZ
 
 | |
7H52
 
 | | Group deposition for crystallographic fragment screening of Coxsackievirus A16 (G-10) 2A protease -- Crystal structure of Coxsackievirus A16 (G-10) 2A protease in complex with Z1203730981 (A71EV2A-x1209) | | Descriptor: | 2-bromo-1H-imidazole, DIMETHYL SULFOXIDE, Protease 2A, ... | | Authors: | Lithgo, R.M, Fairhead, M, Koekemoer, L, Balcomb, B.H, Capkin, E, Chandran, A.V, Golding, M, Godoy, A.S, Aschenbrenner, J.C, Marples, P.G, Ni, X, Thompson, W, Tomlinson, C.W.E, Wild, C, Winokan, M, Xavier, M.-A.E, Fearon, D, von Delft, F. | | Deposit date: | 2024-04-04 | | Release date: | 2024-04-24 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.21 Å) | | Cite: | Crystallographic Fragment Screen of Coxsackievirus A16 2A Protease identifies new opportunities for the development of broad-spectrum anti-enterovirals.
Biorxiv, 2024
|
|
7H50
 
 | | Group deposition for crystallographic fragment screening of Coxsackievirus A16 (G-10) 2A protease -- Crystal structure of Coxsackievirus A16 (G-10) 2A protease in complex with Z1143441220 (A71EV2A-x1169) | | Descriptor: | 1,3-oxazole-4-carbonitrile, DIMETHYL SULFOXIDE, Protease 2A, ... | | Authors: | Lithgo, R.M, Fairhead, M, Koekemoer, L, Balcomb, B.H, Capkin, E, Chandran, A.V, Golding, M, Godoy, A.S, Aschenbrenner, J.C, Marples, P.G, Ni, X, Thompson, W, Tomlinson, C.W.E, Wild, C, Winokan, M, Xavier, M.-A.E, Fearon, D, von Delft, F. | | Deposit date: | 2024-04-04 | | Release date: | 2024-04-24 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.06 Å) | | Cite: | Crystallographic Fragment Screen of Coxsackievirus A16 2A Protease identifies new opportunities for the development of broad-spectrum anti-enterovirals.
Biorxiv, 2024
|
|
8A23
 
 | | Crystal structure of SARS-CoV-2 nsp10/nsp16 methyltransferase in complex with TO383 | | Descriptor: | (2R,3R,4S,5R)-2-[4-azanyl-5-(2-quinolin-3-ylethynyl)pyrrolo[2,3-d]pyrimidin-7-yl]-5-(hydroxymethyl)oxolane-3,4-diol, 2'-O-methyltransferase nsp16, GLYCEROL, ... | | Authors: | Hanigovsky, M, Krafcikova, P, Klima, M, Boura, E. | | Deposit date: | 2022-06-02 | | Release date: | 2023-06-14 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal structure of SARS-CoV-2 nsp10/nsp16 methyltransferase in
complex with TO383
To Be Published
|
|
4Q07
 
 | | Crystal structure of chimeric carbonic anhydrase IX with inhibitor | | Descriptor: | 3-(cyclooctylamino)-2,5,6-trifluoro-4-[(2-hydroxyethyl)sulfonyl]benzenesulfonamide, BICINE, Carbonic anhydrase 2, ... | | Authors: | Smirnov, A, Manakova, E, Grazulis, S. | | Deposit date: | 2014-04-01 | | Release date: | 2015-01-28 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.15 Å) | | Cite: | Discovery and characterization of novel selective inhibitors of carbonic anhydrase IX.
J.Med.Chem., 57, 2014
|
|
4XDI
 
 | | Structure of Chlamydomonas reinhardtii THB1 | | Descriptor: | Chlamydomonas reinhardtii THB1, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Rice, S.L, Boucher, L.E, Schlessman, J.L, Bosch, J, Lecomte, J.T.J. | | Deposit date: | 2014-12-19 | | Release date: | 2015-01-21 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.893 Å) | | Cite: | Structure of Chlamydomonas reinhardtii THB1, a group 1 truncated hemoglobin with a rare histidine-lysine heme ligation.
Acta Crystallogr.,Sect.F, 71, 2015
|
|
6R5M
 
 | | Crystal structure of toxin MT9 from mamba venom | | Descriptor: | ACETYL GROUP, Dendroaspis polylepis MT9, GLYCEROL, ... | | Authors: | Stura, E.A, Tepshi, L, Ciolek, J, Triquigneaux, M, Zoukimian, C, De Waard, M, Beroud, R, Servent, D, Gilles, N, Legrand, P, Ciccone, L. | | Deposit date: | 2019-03-25 | | Release date: | 2020-02-12 | | Last modified: | 2022-05-25 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | MT9, a natural peptide from black mamba venom antagonizes the muscarinic type 2 receptor and reverses the M2R-agonist-induced relaxation in rat and human arteries
Biomed Pharmacother, 150, 2022
|
|
7H6Q
 
 | | Group deposition for crystallographic fragment screening of Chikungunya virus nsP3 macrodomain -- Crystal structure of Chikungunya virus nsP3 macrodomain in complex with Z1148165337 (CHIKV_MacB-x0312) | | Descriptor: | 2-(1H-indazol-1-yl)-N,N-dimethylacetamide, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, CHLORIDE ION, ... | | Authors: | Aschenbrenner, J.C, Fairhead, M, Godoy, A.S, Balcomb, B.H, Capkin, E, Chandran, A.V, Dolci, I, Golding, M, Koekemoer, L, Lithgo, R.M, Marples, P.G, Ni, X, Oliva, G, Thompson, W, Tomlinson, C.W.E, Wild, C, Winokan, M, Xavier, M.-A.E, Fearon, D, von Delft, F. | | Deposit date: | 2024-04-26 | | Release date: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Group deposition for crystallographic fragment screening of Chikungunya virus nsP3 macrodomain
To Be Published
|
|
7H7E
 
 | | Group deposition for crystallographic fragment screening of Chikungunya virus nsP3 macrodomain -- Crystal structure of Chikungunya virus nsP3 macrodomain in complex with Z432057220 (CHIKV_MacB-x0732) | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, CHLORIDE ION, DIMETHYL SULFOXIDE, ... | | Authors: | Aschenbrenner, J.C, Fairhead, M, Godoy, A.S, Balcomb, B.H, Capkin, E, Chandran, A.V, Dolci, I, Golding, M, Koekemoer, L, Lithgo, R.M, Marples, P.G, Ni, X, Oliva, G, Thompson, W, Tomlinson, C.W.E, Wild, C, Winokan, M, Xavier, M.-A.E, Fearon, D, von Delft, F. | | Deposit date: | 2024-04-26 | | Release date: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Group deposition for crystallographic fragment screening of Chikungunya virus nsP3 macrodomain
To Be Published
|
|
7H7P
 
 | | Group deposition for crystallographic fragment screening of Chikungunya virus nsP3 macrodomain -- Crystal structure of Chikungunya virus nsP3 macrodomain in complex with Z32968340 (CHIKV_MacB-x0926) | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, CHLORIDE ION, DIMETHYL SULFOXIDE, ... | | Authors: | Aschenbrenner, J.C, Fairhead, M, Godoy, A.S, Balcomb, B.H, Capkin, E, Chandran, A.V, Dolci, I, Golding, M, Koekemoer, L, Lithgo, R.M, Marples, P.G, Ni, X, Oliva, G, Thompson, W, Tomlinson, C.W.E, Wild, C, Winokan, M, Xavier, M.-A.E, Fearon, D, von Delft, F. | | Deposit date: | 2024-04-26 | | Release date: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (1.43 Å) | | Cite: | Group deposition for crystallographic fragment screening of Chikungunya virus nsP3 macrodomain
To Be Published
|
|
6E4L
 
 | | The structure of the N-terminal domain of human clathrin heavy chain 1 (nTD) in complex with ES9 | | Descriptor: | 1,2-ETHANEDIOL, 5-bromo-N-(4-nitrophenyl)thiophene-2-sulfonamide, ACETATE ION, ... | | Authors: | Dejonghe, W, Sharma, I, Denoo, B, Munck, S.D, Bulut, H, Mylle, E, Vasileva, M, Lu, Q, Savatin, D.V, Mishev, K, Nerinckx, W, Staes, A, Drozdzecki, A, Audenaert, D, Madder, A, Friml, J, Damme, D.V, Gevaert, K, Haucke, V, Savvides, S, Winne, J, Russinova, E. | | Deposit date: | 2018-07-17 | | Release date: | 2019-04-24 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Disruption of endocytosis through chemical inhibition of clathrin heavy chain function.
Nat.Chem.Biol., 15, 2019
|
|
5ITB
 
 | | Crystal structure of the anti-RSV F Fab 14N4 | | Descriptor: | anti-RSV F Fab 14N4 Heavy chain, anti-RSV F Fab 14N4 Light chain | | Authors: | Mousa, J.J, Crowe, J.E. | | Deposit date: | 2016-03-16 | | Release date: | 2016-10-26 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural basis for nonneutralizing antibody competition at antigenic site II of the respiratory syncytial virus fusion protein.
Proc.Natl.Acad.Sci.USA, 113, 2016
|
|
