2OOH
 
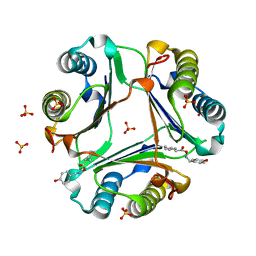 | | Crystal Structure of MIF bound to a Novel Inhibitor, OXIM-11 | | Descriptor: | 4-HYDROXYBENZALDEHYDE O-(CYCLOHEXYLCARBONYL)OXIME, GLYCEROL, Macrophage migration inhibitory factor, ... | | Authors: | Crichlow, G.V, Al-Abed, Y, Lolis, E. | | Deposit date: | 2007-01-25 | | Release date: | 2007-06-05 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Alternative chemical modifications reverse the binding orientation of a pharmacophore scaffold in the active site of macrophage migration inhibitory factor.
J.Biol.Chem., 282, 2007
|
|
2OQ6
 
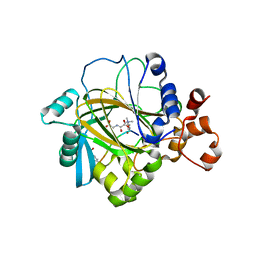 | | Crystal structure of JMJD2A complexed with histone H3 peptide trimethylated at Lys9 | | Descriptor: | JmjC domain-containing histone demethylation protein 3A, N-OXALYLGLYCINE, NICKEL (II) ION, ... | | Authors: | Pilka, E.S, Ng, S.S, Kavanagh, K.L, McDonough, M.A, Savitsky, P, von Delft, F, Arrowsmith, C.H, Weigelt, J, Edwards, A, Sundstrom, M, Schofield, C.J, Oppermann, U, Structural Genomics Consortium (SGC) | | Deposit date: | 2007-01-31 | | Release date: | 2007-03-13 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structures of histone demethylase JMJD2A reveal basis for substrate specificity.
Nature, 448, 2007
|
|
4CYJ
 
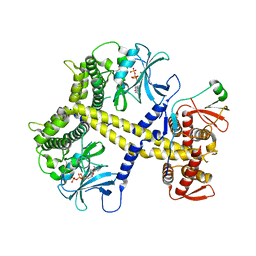 | | Chaetomium thermophilum Pan2:Pan3 complex | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, PAB-DEPENDENT POLY(A)-SPECIFIC RIBONUCLEASE SUBUNIT PAN3-LIKE PROTEIN, ... | | Authors: | Wolf, J, Valkov, E, Allen, M.D, Meineke, B, Gordiyenko, Y, McLaughlin, S.H, Olsen, T.M, Robinson, C.V, Bycroft, M, Stewart, M, Passmore, L.A. | | Deposit date: | 2014-04-11 | | Release date: | 2014-06-11 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.59 Å) | | Cite: | Structural Basis for Pan3 Binding to Pan2 and its Function in Mrna Recruitment and Deadenylation
Embo J., 33, 2014
|
|
4URP
 
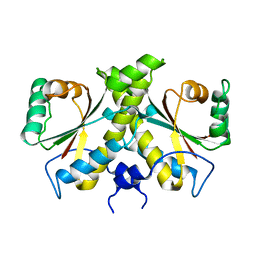 | | The Crystal structure of Nitroreductase from Saccharomyces cerevisiae | | Descriptor: | FATTY ACID REPRESSION MUTANT PROTEIN 2 | | Authors: | Song, H.-N, Woo, E.-J, Bang, S.-Y, Jung, D.-G, Park, S.-G. | | Deposit date: | 2014-07-01 | | Release date: | 2015-04-29 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.991 Å) | | Cite: | Crystal Structure of the Fungal Nitroreductase Frm2 from Saccharomyces Cerevisiae.
Protein Sci., 24, 2015
|
|
1U92
 
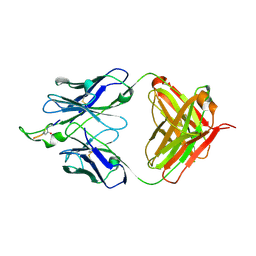 | | Crystal structure of the HIV-1 Cross Neutralizing Monoclonal Antibody 2F5 in complex with gp41 Peptide Analog E-[Dap]-DKWQS (cyclic) | | Descriptor: | ANTIBODY 2F5 (HEAVY CHAIN), ANTIBODY 2F5 (LIGHT CHAIN), GP41 PEPTIDE ANALOG | | Authors: | Bryson, S, Julien, J.-P, Hynes, R.C, Pai, E.F. | | Deposit date: | 2004-08-09 | | Release date: | 2004-10-05 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.24 Å) | | Cite: | Crystallographic definition of the epitope promiscuity of the broadly neutralizing anti-human immunodeficiency virus type 1 antibody 2F5: vaccine design implications
J.Virol., 83, 2009
|
|
1TRA
 
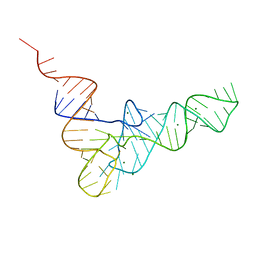 | | RESTRAINED REFINEMENT OF THE MONOCLINIC FORM OF YEAST PHENYLALANINE TRANSFER RNA. TEMPERATURE FACTORS AND DYNAMICS, COORDINATED WATERS, AND BASE-PAIR PROPELLER TWIST ANGLES | | Descriptor: | MAGNESIUM ION, TRNAPHE | | Authors: | Westhof, E, Sundaralingam, M. | | Deposit date: | 1986-05-16 | | Release date: | 1986-07-14 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Restrained refinement of the monoclinic form of yeast phenylalanine transfer RNA. Temperature factors and dynamics, coordinated waters, and base-pair propeller twist angles.
Biochemistry, 25, 1986
|
|
7OE7
 
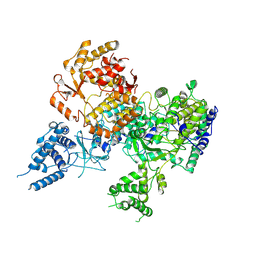 | | Apo-structure of Lassa virus L protein (well-resolved alpha ribbon) [APO-RIBBON] | | Descriptor: | MAGNESIUM ION, RNA-directed RNA polymerase L, ZINC ION | | Authors: | Kouba, T, Vogel, D, Thorkelsson, S, Quemin, E, Williams, H.M, Milewski, M, Busch, C, Gunther, S, Grunewald, K, Rosenthal, M, Cusack, S. | | Deposit date: | 2021-05-01 | | Release date: | 2021-12-01 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (3.73 Å) | | Cite: | Conformational changes in Lassa virus L protein associated with promoter binding and RNA synthesis activity.
Nat Commun, 12, 2021
|
|
2P76
 
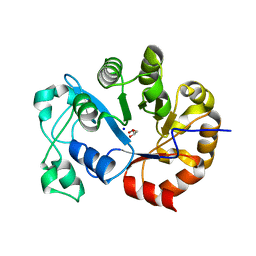 | | Crystal structure of a Glycerophosphodiester Phosphodiesterase from Staphylococcus aureus | | Descriptor: | GLYCEROL, Glycerophosphoryl diester phosphodiesterase, SODIUM ION | | Authors: | Fedorov, A.A, Fedorov, E.V, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2007-03-20 | | Release date: | 2007-04-03 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Crystal structure of a Glycerophosphodiester Phosphodiesterase from Staphylococcus aureus
To be Published
|
|
1KN9
 
 | |
7OE3
 
 | | Apo-structure of Lassa virus L protein (well-resolved endonuclease) [APO-ENDO] | | Descriptor: | MAGNESIUM ION, RNA-directed RNA polymerase L, ZINC ION | | Authors: | Kouba, T, Vogel, D, Thorkelsson, S, Quemin, E, Williams, H.M, Milewski, M, Busch, C, Gunther, S, Grunewald, K, Rosenthal, M, Cusack, S. | | Deposit date: | 2021-05-01 | | Release date: | 2021-12-01 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (3.35 Å) | | Cite: | Conformational changes in Lassa virus L protein associated with promoter binding and RNA synthesis activity.
Nat Commun, 12, 2021
|
|
7OCH
 
 | | Apo-structure of Lassa virus L protein (well-resolved polymerase core) [APO-CORE] | | Descriptor: | MAGNESIUM ION, RNA-directed RNA polymerase L, ZINC ION | | Authors: | Kouba, T, Vogel, D, Thorkelsson, S, Quemin, E, Williams, H.M, Milewski, M, Busch, C, Gunther, S, Grunewald, K, Rosenthal, M, Cusack, S. | | Deposit date: | 2021-04-26 | | Release date: | 2021-12-01 | | Last modified: | 2024-05-01 | | Method: | ELECTRON MICROSCOPY (3.14 Å) | | Cite: | Conformational changes in Lassa virus L protein associated with promoter binding and RNA synthesis activity.
Nat Commun, 12, 2021
|
|
4UZ5
 
 | |
4MCE
 
 | | Crystal structure of the Gas5 GRE Mimic | | Descriptor: | Gas5 GREM Fwd, Gas5 GREM Rev, SULFATE ION | | Authors: | Hudson, W.H, Ortlund, E.A. | | Deposit date: | 2013-08-21 | | Release date: | 2014-09-24 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.205 Å) | | Cite: | Conserved sequence-specific lincRNA-steroid receptor interactions drive transcriptional repression and direct cell fate.
Nat Commun, 5, 2014
|
|
4DKR
 
 | | Crystal structure of clade A/E 93TH057 HIV-1 gp120 core in complex with AWS-I-169 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, HIV-1 gp120 core, ... | | Authors: | Kwon, Y.D, LaLonde, J.M, Jones, D.M, Sun, A.W, Courter, J.R, Soeta, T, Kobayashi, T, Princiotto, A.M, Wu, X, Mascola, J, Schon, A, Freire, E, Sodroski, J, Madani, N, Smith III, A.B, Kwong, P.D. | | Deposit date: | 2012-02-03 | | Release date: | 2012-05-02 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structure-Based Design, Synthesis, and Characterization of Dual Hotspot Small-Molecule HIV-1 Entry Inhibitors.
J.Med.Chem., 55, 2012
|
|
1TUJ
 
 | | Solution structure of the honey bee general odorant binding protein ASP2 in complex with trimethylsilyl-d4 propionate | | Descriptor: | 3-TRIMETHYLSILYL-PROPIONATE-2,2,3,3,-D4, odorant binding protein ASP2 | | Authors: | Lescop, E, Briand, L, Pernollet, J.-C, Guittet, E. | | Deposit date: | 2004-06-25 | | Release date: | 2005-09-20 | | Last modified: | 2024-10-30 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the honey bee general odorant binding protein ASP2 in complex with trimethylsilyl-d4 propionate
To be Published
|
|
1KFT
 
 | | Solution Structure of the C-Terminal domain of UvrC from E-coli | | Descriptor: | Excinuclease ABC subunit C | | Authors: | Singh, S, Folkers, G.E, Bonvin, A.M.J.J, Boelens, R, Wechselberger, R, Niztayev, A, Kaptein, R. | | Deposit date: | 2001-11-23 | | Release date: | 2002-11-20 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Solution structure and DNA-binding properties of the C-terminal domain of UvrC from E.coli
EMBO J., 21, 2002
|
|
1KK3
 
 | | Structure of the wild-type large gamma subunit of initiation factor eIF2 from Pyrococcus abyssi complexed with GDP-Mg2+ | | Descriptor: | GUANOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, ZINC ION, ... | | Authors: | Schmitt, E, Blanquet, S, Mechulam, Y. | | Deposit date: | 2001-12-06 | | Release date: | 2002-04-10 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | The large subunit of initiation factor aIF2 is a close structural homologue of elongation factors.
EMBO J., 21, 2002
|
|
2OZ7
 
 | | Crystal structure of the human androgen receptor T877A mutant ligand-binding domain with cyproterone acetate | | Descriptor: | Androgen receptor, CYPROTERONE ACETATE | | Authors: | Bohl, C.E, Wu, Z, Miller, D.D, Bell, C.E, Dalton, J.T. | | Deposit date: | 2007-02-25 | | Release date: | 2007-03-27 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure of the T877A human androgen receptor ligand-binding domain complexed to cyproterone acetate provides insight for ligand-induced conformational changes and structure-based drug design.
J.Biol.Chem., 282, 2007
|
|
2OQ7
 
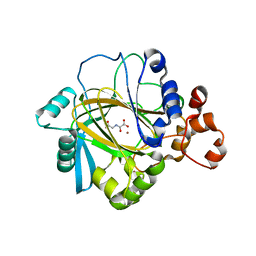 | | The crystal structure of JMJD2A complexed with Ni and N-oxalylglycine | | Descriptor: | JmjC domain-containing histone demethylation protein 3A, N-OXALYLGLYCINE, NICKEL (II) ION, ... | | Authors: | Kavanagh, K.L, Ng, S.S, Pilka, E, McDonough, M.A, Savitsky, P, von Delft, F, Arrowsmith, C.H, Weigelt, J, Edwards, A, Sundstrom, M, Schofield, C.J, Oppermann, U, Structural Genomics Consortium (SGC) | | Deposit date: | 2007-01-31 | | Release date: | 2007-03-13 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Crystal structures of histone demethylase JMJD2A reveal basis for substrate specificity.
Nature, 448, 2007
|
|
1UGH
 
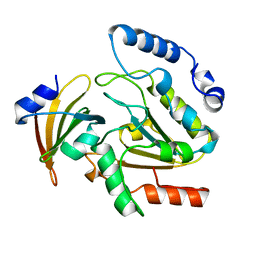 | | CRYSTAL STRUCTURE OF HUMAN URACIL-DNA GLYCOSYLASE IN COMPLEX WITH A PROTEIN INHIBITOR: PROTEIN MIMICRY OF DNA | | Descriptor: | PROTEIN (URACIL-DNA GLYCOSYLASE INHIBITOR), PROTEIN (URACIL-DNA GLYCOSYLASE) | | Authors: | Mol, C.D, Arvai, A.S, Sanderson, R.J, Slupphaug, G, Kavli, B, Krokan, H.E, Mosbaugh, D.W, Tainer, J.A. | | Deposit date: | 1999-02-05 | | Release date: | 1999-02-16 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of human uracil-DNA glycosylase in complex with a protein inhibitor: protein mimicry of DNA.
Cell(Cambridge,Mass.), 82, 1995
|
|
6ZIE
 
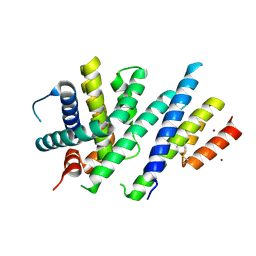 | | Crystal structure of MCL-1 in complex with a neutralizing Alphabody CMPX-383B | | Descriptor: | CMPX-383B, Induced myeloid leukemia cell differentiation protein Mcl-1, ZINC ION | | Authors: | Pannecoucke, E, Savvides, S.N, Desmet, J, Lasters, I. | | Deposit date: | 2020-06-25 | | Release date: | 2021-04-28 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Cell-penetrating Alphabody protein scaffolds for intracellular drug targeting.
Sci Adv, 7, 2021
|
|
2P4M
 
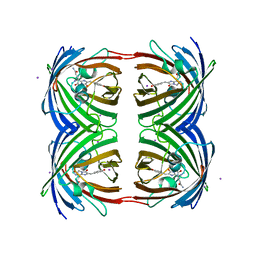 | | High pH structure of Rtms5 H146S variant | | Descriptor: | GFP-like non-fluorescent chromoprotein, IODIDE ION | | Authors: | Battad, J.M, Wilmann, P.G, Olsen, S, Byres, E, Smith, S.C, Dove, S.G, Turcic, K.N, Devenish, R.J, Rossjohn, J, Prescott, M. | | Deposit date: | 2007-03-12 | | Release date: | 2007-04-03 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | A structural basis for the pH-dependent increase in fluorescence efficiency of chromoproteins
J.Mol.Biol., 368, 2007
|
|
6F2I
 
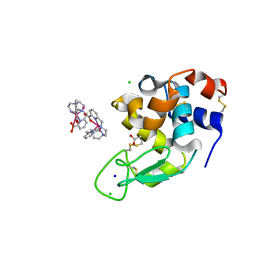 | | Crystal structure of Hen Egg-White Lysozyme co-crystallized in presence of 100 mM Tb-Xo4 | | Descriptor: | ACETATE ION, CHLORIDE ION, Lysozyme C, ... | | Authors: | Engilberge, S, Riobe, F, DI Pietro, S, Girard, E, Dumont, E, Maury, O. | | Deposit date: | 2017-11-24 | | Release date: | 2018-10-03 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Unveiling the Binding Modes of the Crystallophore, a Terbium-based Nucleating and Phasing Molecular Agent for Protein Crystallography.
Chemistry, 24, 2018
|
|
2P6X
 
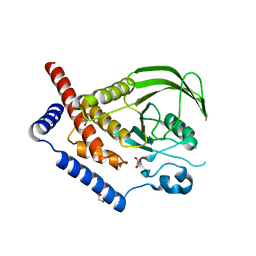 | | Crystal structure of human tyrosine phosphatase PTPN22 | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, Tyrosine-protein phosphatase non-receptor type 22 | | Authors: | Ugochukwu, E, Salah, E, Barr, A, Shrestha, L, Alfano, I, Burgess-Brown, N, King, O, Umeano, C, Papagrigoriou, E, Pike, A.C.W, Bunkoczi, G, Turnbull, A, Uppenberg, J, Sundstrom, M, Arrowsmith, C.H, Weigelt, J, Edwards, A, von Delft, F, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2007-03-19 | | Release date: | 2007-04-03 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Large-scale structural analysis of the classical human protein tyrosine phosphatome.
Cell(Cambridge,Mass.), 136, 2009
|
|
6F2K
 
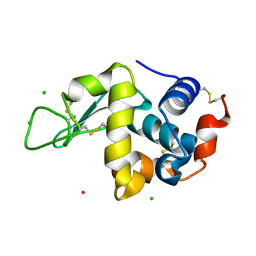 | | Crystal structure of Hen Egg-White Lysozyme co-crystallized in presence of 100 mM Tb-Xo4 and 100 mM potassium phosphate monobasic. | | Descriptor: | CHLORIDE ION, Lysozyme C, TERBIUM(III) ION | | Authors: | Engilberge, S, Riobe, F, Di Pietro, S, Girard, E, Dumont, E, Maury, O. | | Deposit date: | 2017-11-24 | | Release date: | 2018-10-03 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Unveiling the Binding Modes of the Crystallophore, a Terbium-based Nucleating and Phasing Molecular Agent for Protein Crystallography.
Chemistry, 24, 2018
|
|
