4IUR
 
 | |
2I5E
 
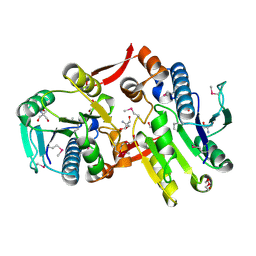 | | Crystal Structure of a Protein of Unknown Function MM2497 from Methanosarcina mazei Go1, Probable Nucleotidyltransferase | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, DI(HYDROXYETHYL)ETHER, Hypothetical protein MM_2497 | | Authors: | Tan, K, Du, J, Clancy, S, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2006-08-24 | | Release date: | 2006-09-26 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | The crystal structure of a hypothetical protein MM_2497 from Methanosarcina mazei Go1
To be Published
|
|
5XO1
 
 | |
5YKO
 
 | |
5YKN
 
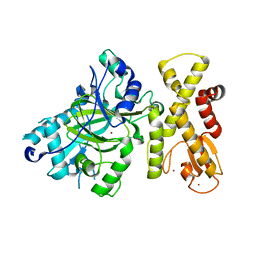 | | crystal structure of Arabidopsis thaliana JMJ14 catalytic domain | | Descriptor: | NICKEL (II) ION, Probable lysine-specific demethylase JMJ14, ZINC ION | | Authors: | Yang, Z, Du, J. | | Deposit date: | 2017-10-15 | | Release date: | 2017-12-27 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structure of the Arabidopsis JMJ14-H3K4me3 Complex Provides Insight into the Substrate Specificity of KDM5 Subfamily Histone Demethylases.
Plant Cell, 30, 2018
|
|
5ZNP
 
 | | Crystal structure of PtSHL in complex with an H3K4me3 peptide | | Descriptor: | 15-mer peptide from Histone H3.2, SHORT LIFE family protein, ZINC ION | | Authors: | Lv, X, Du, J. | | Deposit date: | 2018-04-10 | | Release date: | 2018-07-18 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Dual recognition of H3K4me3 and H3K27me3 by a plant histone reader SHL.
Nat Commun, 9, 2018
|
|
5ZNR
 
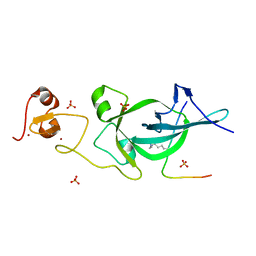 | | Crystal structure of PtSHL in complex with an H3K27me3 peptide | | Descriptor: | 17-mer peptide from Histone H3.2, SHORT LIFE family protein, SULFATE ION, ... | | Authors: | Lv, X, Du, J. | | Deposit date: | 2018-04-10 | | Release date: | 2018-07-18 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (3.202 Å) | | Cite: | Dual recognition of H3K4me3 and H3K27me3 by a plant histone reader SHL.
Nat Commun, 9, 2018
|
|
6A5M
 
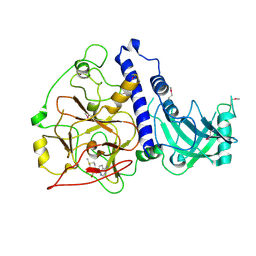 | | Crystal structure of Arabidopsis thaliana SUVH6 in complex with SAM, form 2 | | Descriptor: | Histone-lysine N-methyltransferase, H3 lysine-9 specific SUVH6, S-ADENOSYLMETHIONINE, ... | | Authors: | Li, X, Du, J. | | Deposit date: | 2018-06-24 | | Release date: | 2018-08-29 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.301 Å) | | Cite: | Mechanistic insights into plant SUVH family H3K9 methyltransferases and their binding to context-biased non-CG DNA methylation.
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
6A5N
 
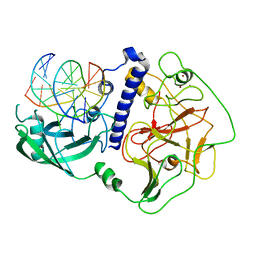 | | Crystal structure of Arabidopsis thaliana SUVH6 in complex with methylated DNA | | Descriptor: | DNA (5'-D(*CP*AP*CP*TP*GP*CP*TP*GP*AP*GP*TP*AP*CP*T)-3'), DNA (5'-D(*GP*AP*GP*TP*AP*CP*TP*(5CM)P*AP*GP*CP*AP*GP*T)-3'), Histone-lysine N-methyltransferase, ... | | Authors: | Li, X, Du, J. | | Deposit date: | 2018-06-24 | | Release date: | 2018-08-29 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Mechanistic insights into plant SUVH family H3K9 methyltransferases and their binding to context-biased non-CG DNA methylation.
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
6A5K
 
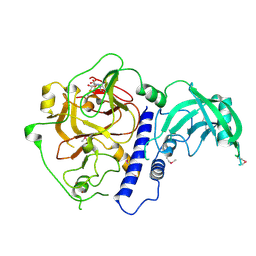 | | Crystal structure of Arabidopsis thaliana SUVH6 in complex with SAM, form 1 | | Descriptor: | Histone-lysine N-methyltransferase, H3 lysine-9 specific SUVH6, S-ADENOSYLMETHIONINE, ... | | Authors: | Li, X, Du, J. | | Deposit date: | 2018-06-24 | | Release date: | 2018-08-29 | | Last modified: | 2018-09-26 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Mechanistic insights into plant SUVH family H3K9 methyltransferases and their binding to context-biased non-CG DNA methylation.
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
3Q7K
 
 | | Formate Channel FocA from Salmonella typhimurium | | Descriptor: | FORMIC ACID, Probable formate transporter | | Authors: | Lue, W, Du, J, Wacker, T, Gerbig-Smentek, E, Andrade, S.L.A, Einsle, O. | | Deposit date: | 2011-01-05 | | Release date: | 2011-04-27 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | pH-dependent gating in a FocA formate channel
Science, 332, 2011
|
|
6MAL
 
 | |
3WMJ
 
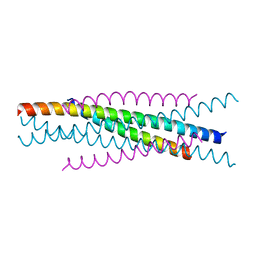 | | Crystal structure of EIAV vaccine gp45 | | Descriptor: | EIAV vaccine gp45 | | Authors: | Liu, X, Du, J, Qiao, W. | | Deposit date: | 2013-11-19 | | Release date: | 2014-11-19 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.998 Å) | | Cite: | A mutation associated with EIAV vaccine strain within heptad repeat of EIAV gp45 provides insight into vaccine development for HIV
To be Published
|
|
3WMI
 
 | | Crystal structure of EIAV wild type gp45 | | Descriptor: | EIAV gp45 wild type | | Authors: | Liu, X, Du, J, Qiao, W. | | Deposit date: | 2013-11-19 | | Release date: | 2014-11-19 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | A mutation associated with EIAV vaccine strain within heptad repeat of EIAV gp45 provides insight into vaccine development for HIV
To be Published
|
|
7Y01
 
 | |
7XPJ
 
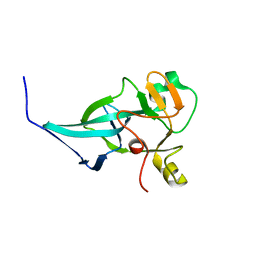 | | crystal structure of rice ASI1 BAH domain | | Descriptor: | BAH domain-containing protein | | Authors: | Yuan, J, Du, J. | | Deposit date: | 2022-05-04 | | Release date: | 2023-01-11 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.301 Å) | | Cite: | Molecular basis of locus-specific H3K9 methylation catalyzed by SUVH6 in plants.
Proc.Natl.Acad.Sci.USA, 120, 2023
|
|
7XPK
 
 | |
5UOW
 
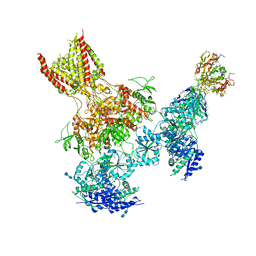 | | Triheteromeric NMDA receptor GluN1/GluN2A/GluN2B in complex with glycine, glutamate, MK-801 and a GluN2B-specific Fab, at pH 6.5 | | Descriptor: | (5S,10R)-5-methyl-10,11-dihydro-5H-5,10-epiminodibenzo[a,d][7]annulene, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, GLUTAMIC ACID, ... | | Authors: | Lu, W, Du, J, Goehring, A, Gouaux, E. | | Deposit date: | 2017-02-01 | | Release date: | 2017-03-22 | | Last modified: | 2020-07-29 | | Method: | ELECTRON MICROSCOPY (4.5 Å) | | Cite: | Cryo-EM structures of the triheteromeric NMDA receptor and its allosteric modulation.
Science, 355, 2017
|
|
5UP2
 
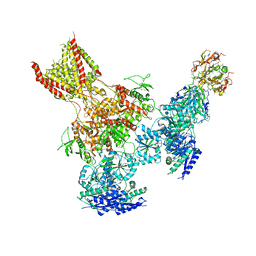 | | Triheteromeric NMDA receptor GluN1/GluN2A/GluN2B in complex with glycine, glutamate, Ro 25-6981, MK-801 and a GluN2B-specific Fab, at pH 6.5 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, GluN2B-specific Fab, ... | | Authors: | Lu, W, Du, J, Goehring, A, Gouaux, E. | | Deposit date: | 2017-02-01 | | Release date: | 2017-03-22 | | Last modified: | 2020-07-29 | | Method: | ELECTRON MICROSCOPY (6 Å) | | Cite: | Cryo-EM structures of the triheteromeric NMDA receptor and its allosteric modulation.
Science, 355, 2017
|
|
5WP6
 
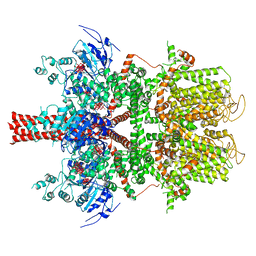 | | Cryo-EM structure of a human TRPM4 channel in complex with calcium and decavanadate | | Descriptor: | DECAVANADATE, Transient receptor potential cation channel subfamily M member 4 | | Authors: | Winkler, P.A, Huang, Y, Sun, W, Du, J, Lu, W. | | Deposit date: | 2017-08-03 | | Release date: | 2017-12-13 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Electron cryo-microscopy structure of a human TRPM4 channel.
Nature, 552, 2017
|
|
6CUD
 
 | | Structure of the human TRPC3 in a lipid-occupied, closed state | | Descriptor: | (2R)-3-hydroxypropane-1,2-diyl dihexanoate, (2S)-3-{[(S)-(2-aminoethoxy)(hydroxy)phosphoryl]oxy}-2-(hexanoyloxy)propyl hexanoate, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Lu, W, Du, J, Fan, C, Choi, W. | | Deposit date: | 2018-03-25 | | Release date: | 2018-05-16 | | Last modified: | 2020-07-29 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Structure of the human lipid-gated cation channel TRPC3.
Elife, 7, 2018
|
|
7EF1
 
 | |
7EF2
 
 | |
7EF3
 
 | |
7EEZ
 
 | | crystal structure of maize SHH2 SAWADEE domain | | Descriptor: | HB transcription factor, ZINC ION | | Authors: | Wang, Y, Du, J. | | Deposit date: | 2021-03-20 | | Release date: | 2021-06-30 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.902 Å) | | Cite: | Recognition of H3K9me1 by maize RNA-directed DNA methylation factor SHH2.
J Integr Plant Biol, 63, 2021
|
|
