2FCT
 
 | | SyrB2 with Fe(II), chloride, and alpha-ketoglutarate | | Descriptor: | ((2R,3S,4S,5S)-3,4-DIHYDROXY-5-(HYDROXYMETHYL)-5-((2R,3S,4S,5S,6R)-3,4,5-TRIHYDROXY-6-METHOXY-TETRAHYDRO-2H-PYRAN-2-YLOXY)-TETRAHYDROFURAN-2-YL)METHYL NONANOATE, 2-OXOGLUTARIC ACID, CHLORIDE ION, ... | | Authors: | Blasiak, L.C, Drennan, C.L. | | Deposit date: | 2005-12-12 | | Release date: | 2006-03-21 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Crystal structure of the non-haem iron halogenase SyrB2 in syringomycin biosynthesis.
Nature, 440, 2006
|
|
2FCU
 
 | | SyrB2 with alpha-ketoglutarate | | Descriptor: | ((2R,3S,4S,5S)-3,4-DIHYDROXY-5-(HYDROXYMETHYL)-5-((2R,3S,4S,5S,6R)-3,4,5-TRIHYDROXY-6-METHOXY-TETRAHYDRO-2H-PYRAN-2-YLOXY)-TETRAHYDROFURAN-2-YL)METHYL NONANOATE, 2-OXOGLUTARIC ACID, syringomycin biosynthesis enzyme 2 | | Authors: | Blasiak, L.C, Drennan, C.L. | | Deposit date: | 2005-12-12 | | Release date: | 2006-03-21 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Crystal structure of the non-haem iron halogenase SyrB2 in syringomycin biosynthesis.
Nature, 440, 2006
|
|
6WTE
 
 | | Structure of radical S-adenosylmethionine methyltransferase, TsrM, from Kitasatospora setae with cobalamin and [4Fe-4S] cluster bound | | Descriptor: | 1,2-ETHANEDIOL, B12-binding domain-containing protein, COBALAMIN, ... | | Authors: | Knox, H.L, Chen, P.Y.-T, Drennan, C.L, Booker, S.J. | | Deposit date: | 2020-05-02 | | Release date: | 2020-12-23 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.67 Å) | | Cite: | Structural basis for non-radical catalysis by TsrM, a radical SAM methylase.
Nat.Chem.Biol., 17, 2021
|
|
6WTF
 
 | | Structure of radical S-adenosylmethionine methyltransferase, TsrM, from Kitasatospora setae with tryptophan substrate and SAM analog (aza-SAM) bound | | Descriptor: | COBALAMIN, IRON/SULFUR CLUSTER, S-5'-AZAMETHIONINE-5'-DEOXYADENOSINE, ... | | Authors: | Knox, H.L, Chen, P.Y.-T, Drennan, C.L, Booker, S.J. | | Deposit date: | 2020-05-02 | | Release date: | 2020-12-23 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.19 Å) | | Cite: | Structural basis for non-radical catalysis by TsrM, a radical SAM methylase.
Nat.Chem.Biol., 17, 2021
|
|
8TWN
 
 | |
8TWT
 
 | |
8TWW
 
 | |
8USP
 
 | |
8USQ
 
 | |
8VHU
 
 | | Crystal structure of dATP bound E. coli class Ia ribonucleotide reductase alpha construct fused with the C-terminal tail of E. coli class Ia beta subunit | | Descriptor: | 2'-DEOXYADENOSINE 5'-TRIPHOSPHATE, CHLORIDE ION, Fusion protein of Ribonucleoside-diphosphate reductase 1 subunits alpha and beta, ... | | Authors: | Funk, M.A, Zimanyi, C.M, Drennan, C.L. | | Deposit date: | 2024-01-02 | | Release date: | 2024-09-04 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | How ATP and dATP Act as Molecular Switches to Regulate Enzymatic Activity in the Prototypical Bacterial Class Ia Ribonucleotide Reductase.
Biochemistry, 63, 2024
|
|
8VHQ
 
 | | Crystal structure of E. coli class Ia ribonucleotide reductase alpha subunit W28A variant bound to dATP and ATP | | Descriptor: | 2'-DEOXYADENOSINE 5'-TRIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Funk, M.A, Zimanyi, C.M, Drennan, C.L. | | Deposit date: | 2024-01-02 | | Release date: | 2024-09-04 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (3.4 Å) | | Cite: | How ATP and dATP Act as Molecular Switches to Regulate Enzymatic Activity in the Prototypical Bacterial Class Ia Ribonucleotide Reductase.
Biochemistry, 63, 2024
|
|
8VHO
 
 | | Crystal Structure of E. coli class Ia ribonucleotide reductase alpha subunit bound to dATP | | Descriptor: | 2'-DEOXYADENOSINE 5'-TRIPHOSPHATE, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, GLYCEROL, ... | | Authors: | Funk, M.A, Zimanyi, C.M, Drennan, C.L. | | Deposit date: | 2024-01-02 | | Release date: | 2024-09-04 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.548 Å) | | Cite: | How ATP and dATP Act as Molecular Switches to Regulate Enzymatic Activity in the Prototypical Bacterial Class Ia Ribonucleotide Reductase.
Biochemistry, 63, 2024
|
|
8VHR
 
 | | Crystal structure of E. coli class Ia ribonucleotide reductase alpha subunit W28A variant bound to dATP and GTP | | Descriptor: | 2'-DEOXYADENOSINE 5'-TRIPHOSPHATE, GUANOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Funk, M.A, Zimanyi, C.M, Drennan, C.L. | | Deposit date: | 2024-01-02 | | Release date: | 2024-09-04 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (3.55 Å) | | Cite: | How ATP and dATP Act as Molecular Switches to Regulate Enzymatic Activity in the Prototypical Bacterial Class Ia Ribonucleotide Reductase.
Biochemistry, 63, 2024
|
|
8VHP
 
 | | Crystal structure of E. coli class Ia ribonucleotide reductase alpha subunit W28A variant bound to CDP and two molecules of ATP | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, CYTIDINE-5'-DIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Funk, M.A, Zimanyi, C.M, Drennan, C.L. | | Deposit date: | 2024-01-02 | | Release date: | 2024-09-04 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.608 Å) | | Cite: | How ATP and dATP Act as Molecular Switches to Regulate Enzymatic Activity in the Prototypical Bacterial Class Ia Ribonucleotide Reductase.
Biochemistry, 63, 2024
|
|
8VHN
 
 | | Crystal Structure of E. coli class Ia ribonucleotide reductase alpha subunit bound to two ATP molecules | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, ADENOSINE-5'-TRIPHOSPHATE, GLYCEROL, ... | | Authors: | Funk, M.A, Zimanyi, C.M, Drennan, C.L. | | Deposit date: | 2024-01-02 | | Release date: | 2024-09-04 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.62 Å) | | Cite: | How ATP and dATP Act as Molecular Switches to Regulate Enzymatic Activity in the Prototypical Bacterial Class Ia Ribonucleotide Reductase.
Biochemistry, 63, 2024
|
|
6XOJ
 
 | |
6XN6
 
 | |
6XPA
 
 | |
6XO3
 
 | | ScoE with alpha-ketoglutarate in an off-site | | Descriptor: | 2-OXOGLUTARIC ACID, CHLORIDE ION, FE (II) ION, ... | | Authors: | Jonnalagadda, R, Drennan, C.L. | | Deposit date: | 2020-07-06 | | Release date: | 2021-01-06 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Biochemical and crystallographic investigations into isonitrile formation by a nonheme iron-dependent oxidase/decarboxylase.
J.Biol.Chem., 296, 2021
|
|
4IXM
 
 | | Crystal structure of Zn(II)-bound YjiA GTPase from E. coli | | Descriptor: | SULFATE ION, Uncharacterized GTP-binding protein YjiA, ZINC ION | | Authors: | Jost, M, Ryan, K.S, Turo, K.E, Drennan, C.L. | | Deposit date: | 2013-01-26 | | Release date: | 2013-02-27 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.57 Å) | | Cite: | Metal binding properties of Escherichia coli YjiA, a member of the metal homeostasis-associated COG0523 family of GTPases.
Biochemistry, 52, 2013
|
|
6MSO
 
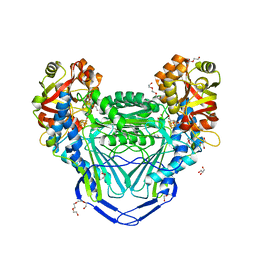 | | Crystal structure of mitochondrial fumarate hydratase from Leishmania major in a complex with inhibitor thiomalate | | Descriptor: | (2S)-2-sulfanylbutanedioic acid, GLYCEROL, IRON/SULFUR CLUSTER, ... | | Authors: | Feliciano, P.R, Drennan, C.L, Nonato, M.C. | | Deposit date: | 2018-10-17 | | Release date: | 2019-01-30 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.053 Å) | | Cite: | Crystal Structures of Fumarate Hydratases from Leishmania major in a Complex with Inhibitor 2-Thiomalate.
ACS Chem. Biol., 14, 2019
|
|
6N2N
 
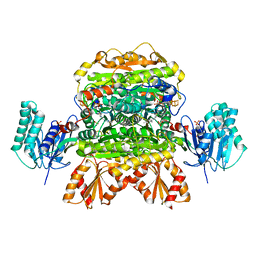 | |
6MSN
 
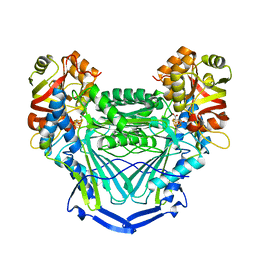 | |
6ND3
 
 | |
4IXN
 
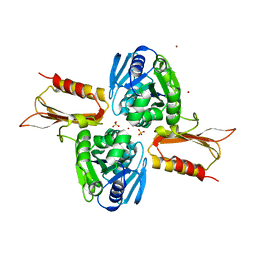 | | Crystal Structure of Zn(II)-bound E37A,C66A,C67A triple mutant YjiA GTPase | | Descriptor: | SULFATE ION, Uncharacterized GTP-binding protein YjiA, ZINC ION | | Authors: | Jost, M, Drennan, C.L. | | Deposit date: | 2013-01-26 | | Release date: | 2013-02-27 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Metal binding properties of Escherichia coli YjiA, a member of the metal homeostasis-associated COG0523 family of GTPases.
Biochemistry, 52, 2013
|
|
