6D51
 
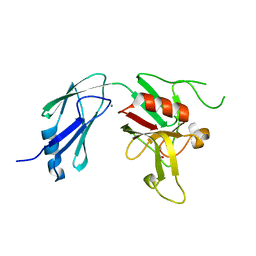 | | Crystal structure of L,D-transpeptidase 3 from Mycobacterium tuberculosis in complex with a faropenem-derived adduct | | Descriptor: | ACETYL GROUP, CALCIUM ION, Probable L,D-transpeptidase 3 | | Authors: | Libreros, G.A, Dias, M.V.B. | | Deposit date: | 2018-04-19 | | Release date: | 2019-02-27 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.83 Å) | | Cite: | Structural Basis for the Interaction and Processing of beta-Lactam Antibiotics by l,d-Transpeptidase 3 (LdtMt3) from Mycobacterium tuberculosis.
ACS Infect Dis, 5, 2019
|
|
8COP
 
 | |
8COQ
 
 | |
8COX
 
 | |
8COW
 
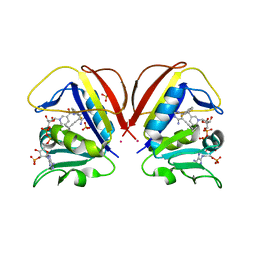 | | Mycobacterium tuberculosis dihydrofolate reductase in complex with 5-(cyclopropylethynyl)-6-(2-fluorophenyl)pyrimidine-2,4-diamine | | Descriptor: | 5-(2-cyclopropylethynyl)-6-(2-fluorophenyl)pyrimidine-2,4-diamine, COBALT (II) ION, Dihydrofolate reductase, ... | | Authors: | Kirkman, T.J, Dias, M.V.B, Coyne, A.G. | | Deposit date: | 2023-03-01 | | Release date: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Currently unpublished
To Be Published
|
|
3KX8
 
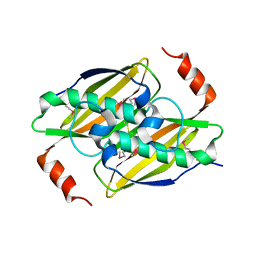 | | Structural basis of the activity and substrate specificity of the fluoroacetyl-CoA thioesterase FlK | | Descriptor: | fluoroacetyl-CoA thioesterase | | Authors: | Chirgadze, D.Y, Dias, M.V.B, Huang, F, Tosin, M, Spiteller, D, Valentine, E.F, Leadlay, P.F, Spencer, J.B, Blundell, T.L. | | Deposit date: | 2009-12-02 | | Release date: | 2010-04-28 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Structural basis for the activity and substrate specificity of fluoroacetyl-CoA thioesterase FlK.
J.Biol.Chem., 285, 2010
|
|
3N59
 
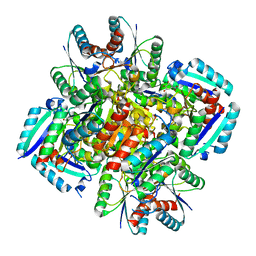 | | Type II dehydroquinase from Mycobacterium Tuberculosis complexed with 3-dehydroshikimate | | Descriptor: | (4S,5R)-4,5-dihydroxy-3-oxocyclohex-1-ene-1-carboxylic acid, 3-dehydroquinate dehydratase, CHLORIDE ION | | Authors: | Snee, W.C, Palaninathan, S.K, Sacchettini, J.C, Dias, M.V.B, Bromfield, K.M, Payne, R, Ciulli, A, Howard, N.I, Abell, C, Blundell, T.L, TB Structural Genomics Consortium (TBSGC) | | Deposit date: | 2010-05-24 | | Release date: | 2010-07-21 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.52 Å) | | Cite: | Structural investigation of inhibitor designs targeting 3-dehydroquinate dehydratase from the shikimate pathway of Mycobacterium tuberculosis.
Biochem.J., 436, 2011
|
|
3N8K
 
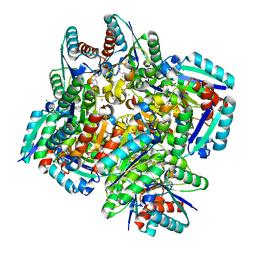 | | Type II dehydroquinase from Mycobacterium tuberculosis complexed with citrazinic acid | | Descriptor: | 2,6-dioxo-1,2,3,6-tetrahydropyridine-4-carboxylic acid, 3-dehydroquinate dehydratase, CHLORIDE ION | | Authors: | Snee, W.C, Palaninathan, S.K, Sacchettini, J.C, Dias, M.V.B, Bromfield, K.M, Payne, R, Ciulli, A, Howard, N.I, Abell, C, Blundell, T.L, TB Structural Genomics Consortium (TBSGC) | | Deposit date: | 2010-05-28 | | Release date: | 2010-07-21 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Structural investigation of inhibitor designs targeting 3-dehydroquinate dehydratase from the shikimate pathway of Mycobacterium tuberculosis.
Biochem.J., 436, 2011
|
|
5U19
 
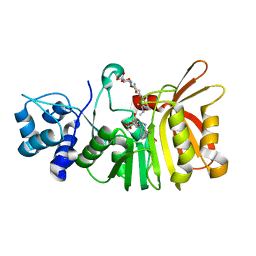 | | Crystal structure of a methyltransferase involved in the biosynthesis of gentamicin in complex with (1R,2S,3S,4R,6R)-4,6-diamino-3-{[3-deoxy-4-C-methyl-3-(methylamino)-beta-L-arabinopyranosyl]oxy}-2-hydroxycyclohexyl 2-amino-2-deoxy-alpha-D-glucopyranoside | | Descriptor: | (1R,2S,3S,4R,6S)-4,6-diamino-3-{[3-deoxy-4-C-methyl-3-(methylamino)-beta-L-arabinopyranosyl]oxy}-2-hydroxycyclohexyl 2-amino-2-deoxy-alpha-D-glucopyranoside, MAGNESIUM ION, Putative gentamicin methyltransferase, ... | | Authors: | Bury, P, Huang, F, Leadlay, P, Dias, M.V.B. | | Deposit date: | 2016-11-28 | | Release date: | 2017-11-01 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.902 Å) | | Cite: | Structural Basis of the Selectivity of GenN, an Aminoglycoside N-Methyltransferase Involved in Gentamicin Biosynthesis.
ACS Chem. Biol., 12, 2017
|
|
5U1I
 
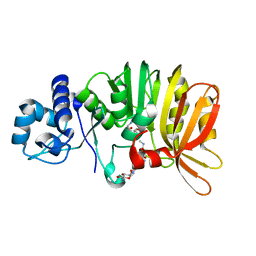 | | Crystal structure of a methyltransferase involved in the biosynthesis of gentamicin in complex with the methylated Kanamycin B | | Descriptor: | (1R,2S,3S,4R,6S)-4,6-diamino-3-{[3-deoxy-3-(methylamino)-alpha-D-glucopyranosyl]oxy}-2-hydroxycyclohexyl 2,6-diamino-2,6-dideoxy-alpha-D-glucopyranoside, CALCIUM ION, Putative gentamicin methyltransferase, ... | | Authors: | Bury, P, Huang, F, Leadlay, P, Dias, M.V.B. | | Deposit date: | 2016-11-28 | | Release date: | 2017-11-01 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.932 Å) | | Cite: | Structural Basis of the Selectivity of GenN, an Aminoglycoside N-Methyltransferase Involved in Gentamicin Biosynthesis.
ACS Chem. Biol., 12, 2017
|
|
5URO
 
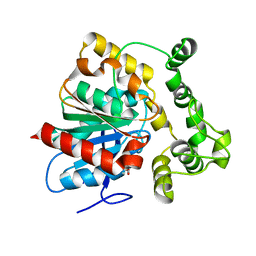 | | Structure of a soluble epoxide hydrolase identified in Trichoderma reesei | | Descriptor: | 2,2'-oxydi(ethyn-1-ol), Predicted protein | | Authors: | Wilson, C, de Oliveira, G, Chambergo, F, Dias, M.V.B. | | Deposit date: | 2017-02-11 | | Release date: | 2017-05-31 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.701 Å) | | Cite: | Structure of a soluble epoxide hydrolase identified in Trichoderma reesei.
Biochim. Biophys. Acta, 1865, 2017
|
|
6NNC
 
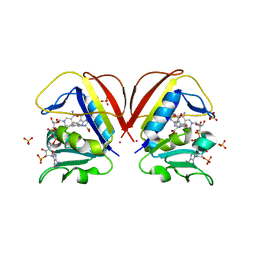 | | Structure of Dihydrofolate reductase from Mycobacterium tuberculosis in complex with NADPH and pemetrexed | | Descriptor: | 2-{4-[2-(2-AMINO-4-OXO-4,7-DIHYDRO-3H-PYRROLO[2,3-D]PYRIMIDIN-5-YL)-ETHYL]-BENZOYLAMINO}-PENTANEDIOIC ACID, COBALT (II) ION, Dihydrofolate reductase, ... | | Authors: | Ribeiro, J.A, Chavez-Pacheco, S.M, Dias, M.V.B. | | Deposit date: | 2019-01-14 | | Release date: | 2019-07-17 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structures of the closed form of Mycobacterium tuberculosis dihydrofolate reductase in complex with dihydrofolate and antifolates.
Acta Crystallogr D Struct Biol, 75, 2019
|
|
6NOI
 
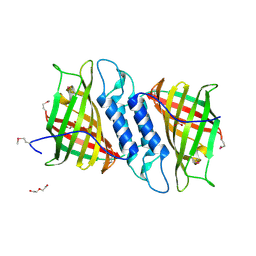 | | Crystal structure of Tsn15 in apo form | | Descriptor: | DI(HYDROXYETHYL)ETHER, PHOSPHATE ION, TRIETHYLENE GLYCOL, ... | | Authors: | Little, R, Paiva, F.C.R, Dias, M.V.B, Leadlay, P. | | Deposit date: | 2019-01-16 | | Release date: | 2019-10-23 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Unexpected enzyme-catalysed [4+2] cycloaddition and rearrangement in polyether antibiotic biosynthesis
Nat Catal, 2019
|
|
6NNW
 
 | | Tsn15 in complex with substrate intermediate | | Descriptor: | (3E)-3-{(1S,4S,4aS,5R,8aS)-1-[(2E,4R,7S,8E,10S)-1,7-dihydroxy-10-{(2R,3S,5R)-5-[(1S)-1-methoxyethyl]-3-methyloxolan-2-yl}-4-methylundeca-2,8-dien-2-yl]-4,5-dimethyloctahydro-3H-2-benzopyran-3-ylidene}oxolane-2,4-dione, PHOSPHATE ION, PROPANOIC ACID, ... | | Authors: | Paiva, F.C.R, Little, R, Leadlay, P, Dias, M.V.B. | | Deposit date: | 2019-01-15 | | Release date: | 2019-10-23 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Unexpected enzyme-catalysed [4+2] cycloaddition and rearrangement in polyether antibiotic biosynthesis
Nat Catal, 2019
|
|
6NNH
 
 | | Structure of Closed state of Dihydrofolate reductase from Mycobacterium tuberculosis in complex with NADPH and cycloguanil | | Descriptor: | 1-(4-chlorophenyl)-6,6-dimethyl-1,6-dihydro-1,3,5-triazine-2,4-diamine, COBALT (II) ION, Dihydrofolate reductase, ... | | Authors: | Giudice, J.H.P, Ribeiro, J.A, Dias, M.V.B. | | Deposit date: | 2019-01-15 | | Release date: | 2019-07-17 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.523 Å) | | Cite: | Crystal structures of the closed form of Mycobacterium tuberculosis dihydrofolate reductase in complex with dihydrofolate and antifolates.
Acta Crystallogr D Struct Biol, 75, 2019
|
|
6NOR
 
 | | Crystal structure of GenD2 from gentamicin A biosynthesis in complex with NAD | | Descriptor: | NICOTINAMIDE-ADENINE-DINUCLEOTIDE, Putative NAD dependent dehydrogenase | | Authors: | Araujo, N.C, Bury, P.S, Huang, F, Leadlay, P.F, Dias, M.V.B. | | Deposit date: | 2019-01-16 | | Release date: | 2019-05-15 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.402 Å) | | Cite: | Crystal Structure of GenD2, an NAD-Dependent Oxidoreductase Involved in the Biosynthesis of Gentamicin.
Acs Chem.Biol., 14, 2019
|
|
6NNI
 
 | | Structure of closed state of Dihydrofolate reductase from Mycobacterium tuberculosis in complex with NADPH and pyrimethamine | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, 5-(4-CHLORO-PHENYL)-6-ETHYL-PYRIMIDINE-2,4-DIAMINE, COBALT (II) ION, ... | | Authors: | Ribeiro, J.A, Chavez-Pacheco, S.M, Dias, M.V.B. | | Deposit date: | 2019-01-15 | | Release date: | 2019-07-17 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.561 Å) | | Cite: | Crystal structures of the closed form of Mycobacterium tuberculosis dihydrofolate reductase in complex with dihydrofolate and antifolates.
Acta Crystallogr D Struct Biol, 75, 2019
|
|
5U0T
 
 | | Crystal structure of a methyltransferase involved in the biosynthesis of gentamicin in complex with (1R,2S,3S,4R,6S)-4,6-diamino-3-{[3-deoxy-3-(methylamino)-alpha-D-glucopyranosyl]oxy}-2-hydroxycyclohexyl 2,6-diamino-2,6-dideoxy-alpha-D-glucopyranoside | | Descriptor: | (1R,2S,3S,4R,6S)-4,6-diamino-3-{[3-deoxy-3-(methylamino)-alpha-D-glucopyranosyl]oxy}-2-hydroxycyclohexyl 2,6-diamino-2,6-dideoxy-alpha-D-glucopyranoside, MAGNESIUM ION, Putative gentamicin methyltransferase, ... | | Authors: | Bury, P, Huang, F, Leadlay, P, Dias, M.V.B. | | Deposit date: | 2016-11-27 | | Release date: | 2017-11-01 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.109 Å) | | Cite: | Structural Basis of the Selectivity of GenN, an Aminoglycoside N-Methyltransferase Involved in Gentamicin Biosynthesis.
ACS Chem. Biol., 12, 2017
|
|
6NND
 
 | | Structure of Dihydrofolate reductase from Mycobacterium tuberculosis in complex with NADPH and dihydrofolate | | Descriptor: | COBALT (II) ION, DIHYDROFOLIC ACID, Dihydrofolate reductase, ... | | Authors: | Ribeiro, J.A, Chavez-Pacheco, S.M, Dias, M.V.B. | | Deposit date: | 2019-01-14 | | Release date: | 2019-07-17 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal structures of the closed form of Mycobacterium tuberculosis dihydrofolate reductase in complex with dihydrofolate and antifolates.
Acta Crystallogr D Struct Biol, 75, 2019
|
|
6NNE
 
 | | Crystal structure of Dihydrofolate reductase from Mycobacterium tuberculosis in complex with diaverdine | | Descriptor: | 5-[(3,4-dimethoxyphenyl)methyl]pyrimidine-2,4-diamine, COBALT (II) ION, Dihydrofolate reductase, ... | | Authors: | Ribeiro, J.A, Chavez-Pacheco, S.M, Dias, M.V.B. | | Deposit date: | 2019-01-14 | | Release date: | 2019-07-17 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.595 Å) | | Cite: | Crystal structures of the closed form of Mycobacterium tuberculosis dihydrofolate reductase in complex with dihydrofolate and antifolates.
Acta Crystallogr D Struct Biol, 75, 2019
|
|
5U18
 
 | | Crystal structure of a methyltransferase involved in the biosynthesis of gentamicin in complex with the Geneticin | | Descriptor: | GENETICIN, N-3'' methyltransferase, S-ADENOSYL-L-HOMOCYSTEINE | | Authors: | Bury, P, Huang, F, Leadlay, P, Dias, M.V.B. | | Deposit date: | 2016-11-28 | | Release date: | 2017-11-01 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.195 Å) | | Cite: | Structural Basis of the Selectivity of GenN, an Aminoglycoside N-Methyltransferase Involved in Gentamicin Biosynthesis.
ACS Chem. Biol., 12, 2017
|
|
5U1E
 
 | | Crystal structure of a methyltransferase involved in the biosynthesis of gentamicin in complex with the kanamycin B | | Descriptor: | (1R,2S,3S,4R,6S)-4,6-DIAMINO-3-[(3-AMINO-3-DEOXY-ALPHA-D-GLUCOPYRANOSYL)OXY]-2-HYDROXYCYCLOHEXYL 2,6-DIAMINO-2,6-DIDEOXY-ALPHA-D-GLUCOPYRANOSIDE, Putative gentamicin methyltransferase, S-ADENOSYL-L-HOMOCYSTEINE | | Authors: | Bury, P, Huang, F, Leadlay, P, Dias, M.V.B. | | Deposit date: | 2016-11-28 | | Release date: | 2017-11-01 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.213 Å) | | Cite: | Structural Basis of the Selectivity of GenN, an Aminoglycoside N-Methyltransferase Involved in Gentamicin Biosynthesis.
ACS Chem. Biol., 12, 2017
|
|
5TYQ
 
 | | Crystal structure of a holoenzyme methyltransferase involved in the biosynthesis of gentamicin | | Descriptor: | MAGNESIUM ION, Putative gentamicin methyltransferase, S-ADENOSYL-L-HOMOCYSTEINE | | Authors: | Bury, P, Huang, F, Leadlay, P, Dias, M.V.B. | | Deposit date: | 2016-11-21 | | Release date: | 2017-11-01 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.163 Å) | | Cite: | Structural Basis of the Selectivity of GenN, an Aminoglycoside N-Methyltransferase Involved in Gentamicin Biosynthesis.
ACS Chem. Biol., 12, 2017
|
|
5U0N
 
 | | Crystal structure of a methyltransferase in complex with the substrate involved in the biosynthesis of gentamicin | | Descriptor: | (1R,2S,3S,4R,6S)-4,6-diamino-3-[(3-amino-3-deoxy-alpha-D-xylopyranosyl)oxy]-2-hydroxycyclohexyl 2-amino-2-deoxy-alpha-D-glucopyranoside, MAGNESIUM ION, Putative gentamicin methyltransferase, ... | | Authors: | Bury, P, Huang, F, Li, S, Sun, Y, Leadlay, P, Dias, M.V.B. | | Deposit date: | 2016-11-25 | | Release date: | 2017-11-01 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.115 Å) | | Cite: | Structural Basis of the Selectivity of GenN, an Aminoglycoside N-Methyltransferase Involved in Gentamicin Biosynthesis.
ACS Chem. Biol., 12, 2017
|
|
5U4T
 
 | | Crystal structure of a methyltransferase involved in the biosynthesis of gentamicin | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, IODIDE ION, Putative gentamicin methyltransferase, ... | | Authors: | Bury, P, Huang, F, Leadlay, P, Dias, M.V.B. | | Deposit date: | 2016-12-06 | | Release date: | 2017-11-01 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.086 Å) | | Cite: | Structural Basis of the Selectivity of GenN, an Aminoglycoside N-Methyltransferase Involved in Gentamicin Biosynthesis.
ACS Chem. Biol., 12, 2017
|
|
