6B0T
 
 | |
8IU6
 
 | | Crystal structure of peptidyl-tRNA hydrolase mutant from Enterococcus faecium | | Descriptor: | GLYCEROL, Peptidyl-tRNA hydrolase | | Authors: | Pandey, R, Tripathi, S, Lanka, A.K, Zohib, M, Pal, R.K, Arora, A. | | Deposit date: | 2023-03-23 | | Release date: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Crystal structure of peptidyl-tRNA hydrolase mutant from Enterococcus faecium
To Be Published
|
|
7Y52
 
 | | Crystal structure of peptidyl-tRNA hydrolase from Enterococcus faecium | | Descriptor: | 1,2-ETHANEDIOL, DI(HYDROXYETHYL)ETHER, Peptidyl-tRNA hydrolase, ... | | Authors: | Pandey, R, Zohib, M, Mundra, S, Pal, R.K, Arora, A. | | Deposit date: | 2022-06-16 | | Release date: | 2024-01-17 | | Last modified: | 2024-08-07 | | Method: | X-RAY DIFFRACTION (1.92 Å) | | Cite: | Characterization of structure of peptidyl-tRNA hydrolase from Enterococcus faecium and its inhibition by a pyrrolinone compound.
Int.J.Biol.Macromol., 275, 2024
|
|
7KC5
 
 | | X-ray structure of Lfa-1 I domain in complex with BMS-68852 collected at 273 K | | Descriptor: | 6-[(5S,9R)-9-(4-cyanophenyl)-3-(3,5-dichlorophenyl)-1-methyl-2,4-dioxo-1,3,7-triazaspiro[4.4]non-7-yl]pyridine-3-carboxylic acid, Integrin alpha-L, MAGNESIUM ION | | Authors: | Woldeyes, R.A, Hallenbeck, K.K, Pfaff, S.J, Lee, G, Cortez, S.V, Kelly, M.J, Akassoglou, K, Arkin, M.R, Fraser, J.S. | | Deposit date: | 2020-10-05 | | Release date: | 2021-10-06 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.86 Å) | | Cite: | Divergent conformational dynamics controls allosteric ligand accessibility across evolutionarily related I-domain-containing integrins
To Be Published
|
|
7KC3
 
 | | X-ray structure of Lfa-1 I domain collected at 273 K | | Descriptor: | Integrin alpha-L, MAGNESIUM ION | | Authors: | Woldeyes, R.A, Hallenbeck, K.K, Pfaff, S.J, Lee, G, Cortez, S.V, Kelly, M.J, Akassoglou, K, Arkin, M.R, Fraser, J.S. | | Deposit date: | 2020-10-05 | | Release date: | 2021-10-06 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Divergent conformational dynamics controls allosteric ligand accessibility across evolutionarily related I-domain-containing integrins
To Be Published
|
|
7KC6
 
 | | X-ray structure of Lfa-1 I domain in complex with Lovastatin collected at 273 K | | Descriptor: | Integrin alpha-L, LOVASTATIN, MAGNESIUM ION | | Authors: | Woldeyes, R.A, Hallenbeck, K.K, Pfaff, S.J, Lee, G, Cortez, S.V, Kelly, M.J, Akassoglou, K, Arkin, M.R, Fraser, J.S. | | Deposit date: | 2020-10-05 | | Release date: | 2021-10-06 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Divergent conformational dynamics controls allosteric ligand accessibility across evolutionarily related I-domain-containing integrins
To Be Published
|
|
3KMD
 
 | |
8U5M
 
 | | Structure of Sts-1 HP domain with rebamipide | | Descriptor: | Rebamipide, Ubiquitin-associated and SH3 domain-containing protein B | | Authors: | Azia, F, Dey, R, French, J.B. | | Deposit date: | 2023-09-12 | | Release date: | 2024-02-07 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.46 Å) | | Cite: | Rebamipide and Derivatives are Potent, Selective Inhibitors of Histidine Phosphatase Activity of the Suppressor of T Cell Receptor Signaling Proteins.
J.Med.Chem., 67, 2024
|
|
7JJO
 
 | | Structural Basis of the Activation of Heterotrimeric Gs-protein by Isoproterenol-bound Beta1-Adrenergic Receptor | | Descriptor: | Beta1-Adrenergic Receptor, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | Authors: | Su, M, Zhu, L, Zhang, Y, Paknejad, N, Dey, R, Huang, J, Lee, M.Y, Williams, D, Jordan, K.D, Eng, E.T, Ernst, O.P, Meyerson, J.R, Hite, R.K, Walz, T, Liu, W, Huang, X.Y. | | Deposit date: | 2020-07-27 | | Release date: | 2020-09-02 | | Last modified: | 2020-10-14 | | Method: | ELECTRON MICROSCOPY (2.6 Å) | | Cite: | Structural Basis of the Activation of Heterotrimeric Gs-Protein by Isoproterenol-Bound beta 1 -Adrenergic Receptor.
Mol.Cell, 80, 2020
|
|
8U7E
 
 | | Structure of Sts-1 HP domain with rebamipide derivative | | Descriptor: | N-(4-ethylbenzoyl)-3-(2-oxo-1,2-dihydroquinolin-4-yl)-L-alanine, Ubiquitin-associated and SH3 domain-containing protein B | | Authors: | Aziz, F, Dey, R, French, J.B. | | Deposit date: | 2023-09-15 | | Release date: | 2024-02-07 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.63 Å) | | Cite: | Rebamipide and Derivatives are Potent, Selective Inhibitors of Histidine Phosphatase Activity of the Suppressor of T Cell Receptor Signaling Proteins.
J.Med.Chem., 67, 2024
|
|
2FP2
 
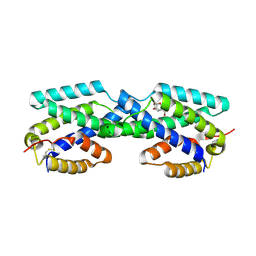 | | Secreted Chorismate Mutase from Mycobacterium tuberculosis | | Descriptor: | 8-HYDROXY-2-OXA-BICYCLO[3.3.1]NON-6-ENE-3,5-DICARBOXYLIC ACID, Chorismate mutase | | Authors: | Okvist, M, Dey, R, Sasso, S, Grahn, E, Kast, P, Krengel, U. | | Deposit date: | 2006-01-15 | | Release date: | 2006-03-28 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.64 Å) | | Cite: | 1.6A Crystal Structure of the Secreted Chorismate Mutase from Mycobacterium tuberculosis: Novel Fold Topology Revealed
J.Mol.Biol., 357, 2006
|
|
2FP1
 
 | | Secreted Chorismate Mutase from Mycobacterium tuberculosis | | Descriptor: | Chorismate mutase, LEAD (II) ION | | Authors: | Okvist, M, Dey, R, Sasso, S, Grahn, E, Kast, P, Krengel, U. | | Deposit date: | 2006-01-15 | | Release date: | 2006-03-28 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | 1.6A Crystal Structure of the Secreted Chorismate Mutase from Mycobacterium tuberculosis: Novel Fold Topology Revealed
J.Mol.Biol., 357, 2006
|
|
7XUZ
 
 | | Crystal structure of a HDAC4-MEF2A-DNA ternary complex | | Descriptor: | DNA (5'-D(*TP*CP*TP*TP*AP*TP*AP*AP*AP*TP*AP*GP*T)-3'), DNA (5'-D(P*AP*CP*TP*AP*TP*TP*TP*AP*TP*AP*A)-3'), Histone deacetylase 4, ... | | Authors: | Dai, S.Y, Guo, L, Dey, R, Guo, M, Bates, D, Cayford, J, Chen, X.J, Wei, X.D, Chen, L, Chen, Y.H. | | Deposit date: | 2022-05-20 | | Release date: | 2023-11-29 | | Last modified: | 2024-06-12 | | Method: | X-RAY DIFFRACTION (3.591 Å) | | Cite: | Structural insights into the HDAC4-MEF2A-DNA complex and its implication in long-range transcriptional regulation.
Nucleic Acids Res., 52, 2024
|
|
4HCA
 
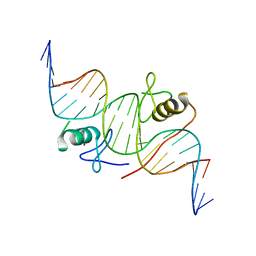 | | DNA binding by GATA transcription factor-complex 1 | | Descriptor: | DNA (5'-D(*AP*AP*TP*GP*TP*CP*CP*AP*TP*CP*TP*GP*AP*TP*AP*AP*GP*AP*CP*G)-3'), DNA (5'-D(*TP*TP*CP*GP*TP*CP*TP*TP*AP*TP*CP*AP*GP*AP*TP*GP*GP*AP*CP*A)-3'), Trans-acting T-cell-specific transcription factor GATA-3, ... | | Authors: | Chen, Y, Bates, D.L, Dey, R, Chen, L. | | Deposit date: | 2012-09-28 | | Release date: | 2012-12-05 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | DNA Binding by GATA Transcription Factor Suggests Mechanisms of DNA Looping and Long-Range Gene Regulation.
Cell Rep, 2, 2012
|
|
4HC7
 
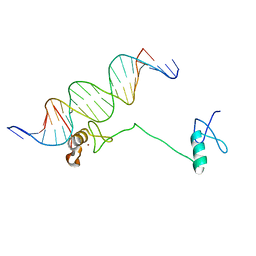 | | Crystal structure of the full DNA binding domain of GATA3-complex 2 | | Descriptor: | DNA (5'-D(*AP*AP*GP*GP*TP*TP*AP*TP*CP*TP*CP*TP*GP*AP*TP*TP*TP*AP*GP*G)-3'), DNA (5'-D(*TP*TP*CP*CP*TP*AP*AP*AP*TP*CP*AP*GP*AP*GP*AP*TP*AP*AP*CP*C)-3'), Trans-acting T-cell-specific transcription factor GATA-3, ... | | Authors: | Chen, Y, Bates, D.L, Dey, R, Chen, L. | | Deposit date: | 2012-09-28 | | Release date: | 2012-12-05 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | DNA Binding by GATA Transcription Factor Suggests Mechanisms of DNA Looping and Long-Range Gene Regulation.
Cell Rep, 2, 2012
|
|
4HC9
 
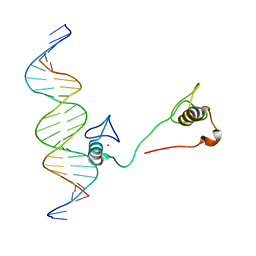 | | DNA binding by GATA transcription factor-complex 3 | | Descriptor: | DNA (5'-D(*AP*AP*GP*GP*TP*TP*AP*TP*CP*TP*CP*TP*GP*AP*TP*TP*TP*AP*GP*G)-3'), DNA (5'-D(*TP*TP*CP*CP*TP*AP*AP*AP*TP*CP*AP*GP*AP*GP*AP*TP*AP*AP*CP*C)-3'), Trans-acting T-cell-specific transcription factor GATA-3, ... | | Authors: | Chen, Y, Bates, D.L, Dey, R, Chen, L. | | Deposit date: | 2012-09-28 | | Release date: | 2012-12-05 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | DNA Binding by GATA Transcription Factor Suggests Mechanisms of DNA Looping and Long-Range Gene Regulation.
Cell Rep, 2, 2012
|
|
3KOV
 
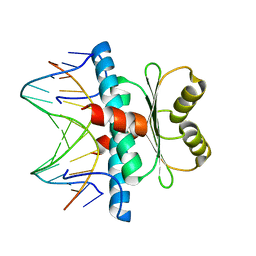 | | Structure of MEF2A bound to DNA reveals a completely folded MADS-box/MEF2 domain that recognizes DNA and recruits transcription co-factors | | Descriptor: | DNA (5'-D(*AP*AP*CP*TP*AP*TP*TP*TP*AP*TP*AP*AP*G)-3'), DNA (5'-D(*TP*CP*TP*TP*AP*TP*AP*AP*AP*TP*AP*GP*T)-3'), Myocyte-specific enhancer factor 2A | | Authors: | Wu, Y, Dey, R, Han, A, Jayathilaka, N, Philips, M, Ye, J, Chen, L. | | Deposit date: | 2009-11-14 | | Release date: | 2010-02-16 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structure of the MADS-box/MEF2 Domain of MEF2A Bound to DNA and Its Implication for Myocardin Recruitment.
J.Mol.Biol., 397, 2010
|
|
3MU6
 
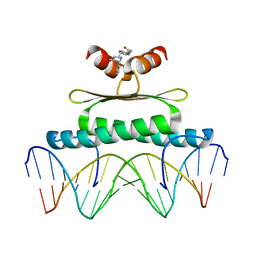 | | Inhibiting the Binding of Class IIa Histone Deacetylases to Myocyte Enhancer Factor-2 by Small Molecules | | Descriptor: | (3E)-N~8~-(2-aminophenyl)-N~1~-phenyloct-3-enediamide, DNA (5'-D(*AP*AP*AP*GP*CP*TP*AP*TP*TP*AP*TP*TP*AP*GP*CP*TP*T)-3'), DNA (5'-D(*TP*AP*AP*GP*CP*TP*AP*AP*TP*AP*AP*TP*AP*GP*CP*TP*T)-3'), ... | | Authors: | Jayathilaka, N, Han, A, Gaffney, K, Dey, R, He, J, Ye, J, Gao, T, Petasis, N.A, Chen, L. | | Deposit date: | 2010-05-01 | | Release date: | 2011-11-02 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.434 Å) | | Cite: | Inhibition of the function of class IIa HDACs by blocking their interaction with MEF2.
Nucleic Acids Res., 40, 2012
|
|
5JOO
 
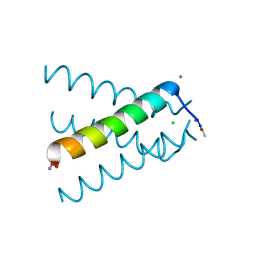 | | XFEL structure of influenza A M2 wild type TM domain at low pH in the lipidic cubic phase at room temperature | | Descriptor: | CALCIUM ION, CHLORIDE ION, Matrix protein 2 | | Authors: | Thomaston, J.L, Woldeyes, R.A, Fraser, J.S, DeGrado, W.F. | | Deposit date: | 2016-05-02 | | Release date: | 2017-08-02 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.413 Å) | | Cite: | XFEL structures of the influenza M2 proton channel: Room temperature water networks and insights into proton conduction.
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
5TTC
 
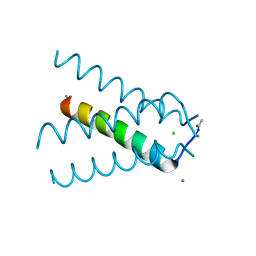 | | XFEL structure of influenza A M2 wild type TM domain at high pH in the lipidic cubic phase at room temperature | | Descriptor: | CALCIUM ION, CHLORIDE ION, Matrix protein 2 | | Authors: | Thomaston, J.L, Woldeyes, R.A, Fraser, J.S, DeGrado, W.F. | | Deposit date: | 2016-11-02 | | Release date: | 2017-08-23 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | XFEL structures of the influenza M2 proton channel: Room temperature water networks and insights into proton conduction.
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
5UM1
 
 | | XFEL structure of influenza A M2 wild type TM domain at intermediate pH in the lipidic cubic phase at room temperature | | Descriptor: | CALCIUM ION, CHLORIDE ION, Matrix protein 2 | | Authors: | Thomaston, J.L, Woldeyes, R.A, Fraser, J.S, DeGrado, W.F. | | Deposit date: | 2017-01-25 | | Release date: | 2017-08-23 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.451 Å) | | Cite: | XFEL structures of the influenza M2 proton channel: Room temperature water networks and insights into proton conduction.
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
4YUH
 
 | | Multiconformer synchrotron model of CypA at 150 K | | Descriptor: | Peptidyl-prolyl cis-trans isomerase A | | Authors: | Keedy, D.A, Kenner, L.R, Warkentin, M, Woldeyes, R.A, Thompson, M.C, Brewster, A.S, Van Benschoten, A.H, Baxter, E.L, Hopkins, J.B, Uervirojnangkoorn, M, McPhillips, S.E, Song, J, Mori, R.A, Holton, J.M, Weis, W.I, Brunger, A.T, Soltis, M, Lemke, H, Gonzalez, A, Sauter, N.K, Cohen, A.E, van den Bedem, H, Thorne, R.E, Fraser, J.S. | | Deposit date: | 2015-03-18 | | Release date: | 2015-10-14 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.34 Å) | | Cite: | Mapping the conformational landscape of a dynamic enzyme by multitemperature and XFEL crystallography.
Elife, 4, 2015
|
|
4YUO
 
 | | High-resolution multiconformer synchrotron model of CypA at 273 K | | Descriptor: | Peptidyl-prolyl cis-trans isomerase A | | Authors: | Keedy, D.A, Kenner, L.R, Warkentin, M, Woldeyes, R.A, Thompson, M.C, Brewster, A.S, Van Benschoten, A.H, Baxter, E.L, Hopkins, J.B, Uervirojnangkoorn, M, McPhillips, S.E, Song, J, Mori, R.A, Holton, J.M, Weis, W.I, Brunger, A.T, Soltis, M, Lemke, H, Gonzalez, A, Sauter, N.K, Cohen, A.E, van den Bedem, H, Thorne, R.E, Fraser, J.S. | | Deposit date: | 2015-03-18 | | Release date: | 2015-05-20 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Mapping the conformational landscape of a dynamic enzyme by multitemperature and XFEL crystallography.
Elife, 4, 2015
|
|
4YUJ
 
 | | Multiconformer synchrotron model of CypA at 240 K | | Descriptor: | Peptidyl-prolyl cis-trans isomerase A | | Authors: | Keedy, D.A, Kenner, L.R, Warkentin, M, Woldeyes, R.A, Thompson, M.C, Brewster, A.S, Van Benschoten, A.H, Baxter, E.L, Hopkins, J.B, Uervirojnangkoorn, M, McPhillips, S.E, Song, J, Mori, R.A, Holton, J.M, Weis, W.I, Brunger, A.T, Soltis, M, Lemke, H, Gonzalez, A, Sauter, N.K, Cohen, A.E, van den Bedem, H, Thorne, R.E, Fraser, J.S. | | Deposit date: | 2015-03-18 | | Release date: | 2015-10-14 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.42 Å) | | Cite: | Mapping the conformational landscape of a dynamic enzyme by multitemperature and XFEL crystallography.
Elife, 4, 2015
|
|
4YUN
 
 | | Multiconformer synchrotron model of CypA at 310 K | | Descriptor: | Peptidyl-prolyl cis-trans isomerase A | | Authors: | Keedy, D.A, Kenner, L.R, Warkentin, M, Woldeyes, R.A, Thompson, M.C, Brewster, A.S, Van Benschoten, A.H, Baxter, E.L, Hopkins, J.B, Uervirojnangkoorn, M, McPhillips, S.E, Song, J, Mori, R.A, Holton, J.M, Weis, W.I, Brunger, A.T, Soltis, M, Lemke, H, Gonzalez, A, Sauter, N.K, Cohen, A.E, van den Bedem, H, Thorne, R.E, Fraser, J.S. | | Deposit date: | 2015-03-18 | | Release date: | 2015-10-14 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.58 Å) | | Cite: | Mapping the conformational landscape of a dynamic enzyme by multitemperature and XFEL crystallography.
Elife, 4, 2015
|
|
