8DMR
 
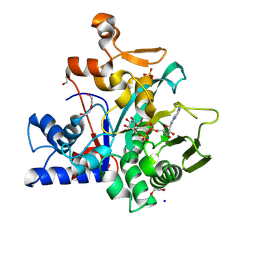 | |
8DMU
 
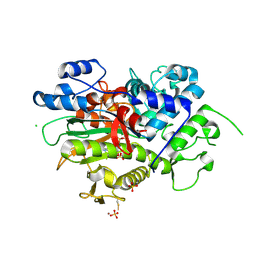 | |
8DMP
 
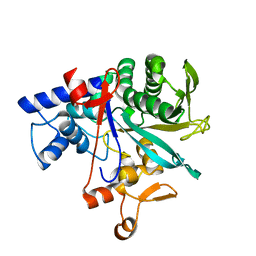 | |
8DMS
 
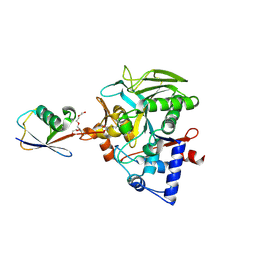 | |
8DMQ
 
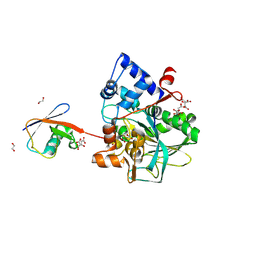 | |
8DMT
 
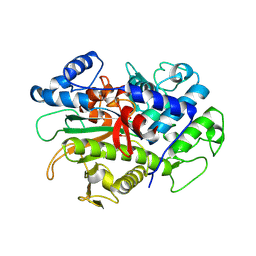 | | Crystal structure of macrodomain CG2909 from Drosophila melanogaster in complex with ADP-ribose | | Descriptor: | RE54994p, [(2R,3S,4R,5R)-5-(6-AMINOPURIN-9-YL)-3,4-DIHYDROXY-OXOLAN-2-YL]METHYL [HYDROXY-[[(2R,3S,4R,5S)-3,4,5-TRIHYDROXYOXOLAN-2-YL]METHOXY]PHOSPHORYL] HYDROGEN PHOSPHATE | | Authors: | Zhang, Z, Das, C. | | Deposit date: | 2022-07-08 | | Release date: | 2023-08-02 | | Last modified: | 2024-07-24 | | Method: | X-RAY DIFFRACTION (2.28 Å) | | Cite: | Legionella metaeffector MavL reverses ubiquitin ADP-ribosylation via a conserved arginine-specific macrodomain.
Nat Commun, 15, 2024
|
|
8DQF
 
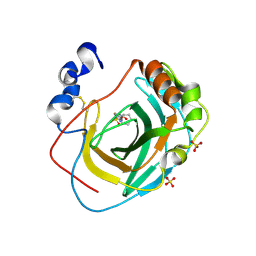 | | Crystal structure of Neisseria gonorrhoeae carbonic anhydrase with N-(5-sulfamoyl-1,3,4-thiadiazol-2-yl)cyclohexanecarboxamide | | Descriptor: | Carbonic anhydrase, N-(5-sulfamoyl-1,3,4-thiadiazol-2-yl)cyclohexanecarboxamide, SULFATE ION, ... | | Authors: | Marapaka, A.K, Das, C, Flaherty, D.P, Yadav, R. | | Deposit date: | 2022-07-19 | | Release date: | 2022-12-14 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structural Characterization of Thiadiazolesulfonamide Inhibitors Bound to Neisseria gonorrhoeae alpha-Carbonic Anhydrase.
Acs Med.Chem.Lett., 14, 2023
|
|
8DYQ
 
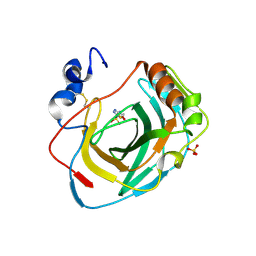 | | Crystal structure of Neisseria gonorrhoeae carbonic anhydrase with Acetazolamide | | Descriptor: | 5-ACETAMIDO-1,3,4-THIADIAZOLE-2-SULFONAMIDE, Carbonic anhydrase, SULFATE ION, ... | | Authors: | Marapaka, A.K, Das, C, Flaherty, D.P, Yadav, R. | | Deposit date: | 2022-08-04 | | Release date: | 2022-12-14 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Structural Characterization of Thiadiazolesulfonamide Inhibitors Bound to Neisseria gonorrhoeae alpha-Carbonic Anhydrase.
Acs Med.Chem.Lett., 14, 2023
|
|
8DR2
 
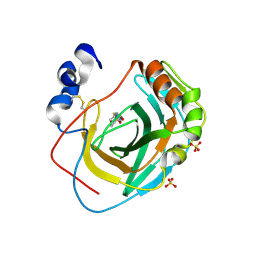 | | Crystal structure of Neisseria gonorrhoeae carbonic anhydrase with 2-cyclohexyl-N-(5-sulfamoyl-1,3,4-thiadiazol-2-yl)acetamide | | Descriptor: | 2-cyclohexyl-N-(5-sulfamoyl-1,3,4-thiadiazol-2-yl)acetamide, Carbonic anhydrase, SULFATE ION, ... | | Authors: | Marapaka, A.K, Das, C, Flaherty, D.P, Yadav, R. | | Deposit date: | 2022-07-20 | | Release date: | 2022-12-14 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.81 Å) | | Cite: | Structural Characterization of Thiadiazolesulfonamide Inhibitors Bound to Neisseria gonorrhoeae alpha-Carbonic Anhydrase.
Acs Med.Chem.Lett., 14, 2023
|
|
8DRB
 
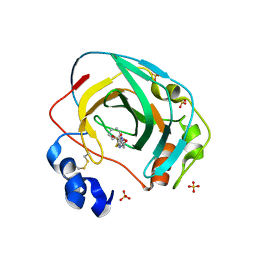 | | Crystal structure of Neisseria gonorrhoeae carbonic anhydrase with 3-phenyl-N-(5-sulfamoyl-1,3,4-thiadiazol-2-yl)propanamide | | Descriptor: | 3-phenyl-N-(5-sulfamoyl-1,3,4-thiadiazol-2-yl)propanamide, Carbonic anhydrase, SULFATE ION, ... | | Authors: | Marapaka, A.K, Das, C, Flaherty, D.P, Yadav, R. | | Deposit date: | 2022-07-20 | | Release date: | 2022-12-14 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.59 Å) | | Cite: | Structural Characterization of Thiadiazolesulfonamide Inhibitors Bound to Neisseria gonorrhoeae alpha-Carbonic Anhydrase.
Acs Med.Chem.Lett., 14, 2023
|
|
8DPC
 
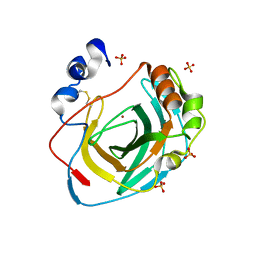 | | Crystal structure of carbonic anhydrase from Neisseria gonorrhoeae | | Descriptor: | Carbonic anhydrase, SULFATE ION, ZINC ION | | Authors: | Marapaka, A.K, Das, C, Flaherty, D.P, Yadav, R. | | Deposit date: | 2022-07-15 | | Release date: | 2022-12-14 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.41 Å) | | Cite: | Structural Characterization of Thiadiazolesulfonamide Inhibitors Bound to Neisseria gonorrhoeae alpha-Carbonic Anhydrase.
Acs Med.Chem.Lett., 14, 2023
|
|
8DPO
 
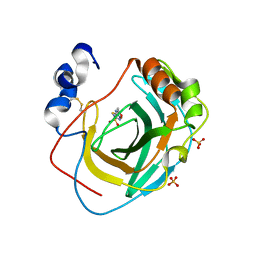 | | Crystal structure of Neisseria gonorrhoeae carbonic anhydrase with Acetazolamide | | Descriptor: | 5-ACETAMIDO-1,3,4-THIADIAZOLE-2-SULFONAMIDE, Carbonic anhydrase, SULFATE ION, ... | | Authors: | Marapaka, A.K, Das, C, Flaherty, D.P, Yadav, R. | | Deposit date: | 2022-07-15 | | Release date: | 2023-07-19 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Crystal structure of Neisseria gonorrhoeae carbonic anhydrase with Acetazolamide
To Be Published
|
|
8EDE
 
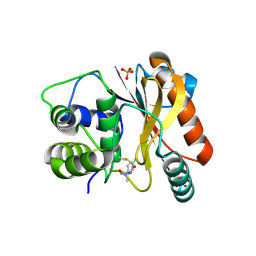 | | Crystal structure of covalent inhibitor 2-chloro-N'-(N-(4-chlorophenyl)-N-methylglycyl)acetohydrazide bound to Ubiquitin C-terminal Hydrolase-L1 | | Descriptor: | 2-[(4-chlorophenyl)-methyl-amino]-~{N}'-ethanoyl-ethanehydrazide, SULFATE ION, Ubiquitin carboxyl-terminal hydrolase isozyme L1 | | Authors: | Patel, R, Imhoff, R, Flaherty, D, Das, C. | | Deposit date: | 2022-09-04 | | Release date: | 2023-09-20 | | Last modified: | 2024-04-10 | | Method: | X-RAY DIFFRACTION (1.799 Å) | | Cite: | Covalent Fragment Screening and Optimization Identifies the Chloroacetohydrazide Scaffold as Inhibitors for Ubiquitin C-terminal Hydrolase L1.
J.Med.Chem., 67, 2024
|
|
8EFW
 
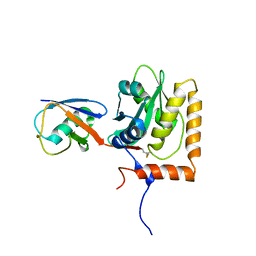 | |
8EFX
 
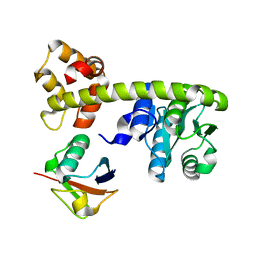 | |
4M3C
 
 | | Structure of a binary complex between homologous tetrameric legume lectins from Butea monosperma and Spatholobus parviflorus seeds | | Descriptor: | CALCIUM ION, GAMMA-AMINO-BUTANOIC ACID, GLYCEROL, ... | | Authors: | Surya, S, Abhilash, J, Geethanandan, K, Sadasivan, C, Haridas, M. | | Deposit date: | 2013-08-06 | | Release date: | 2013-09-18 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structure of a binary complex between homologous tetrameric legume lectins from Butea monosperma and Spatholobus parviflorus seeds
To be Published
|
|
5UBW
 
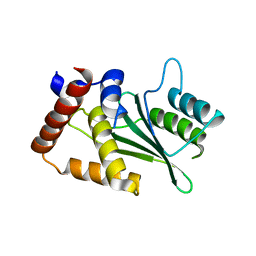 | |
6MRN
 
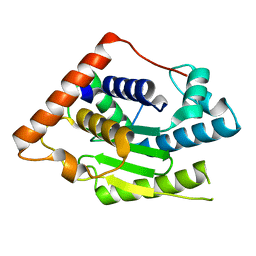 | | Crystal Structure of ChlaDUB2 DUB domain | | Descriptor: | Deubiquitinase and deneddylase Dub2 | | Authors: | Hausman, J.M, Das, C. | | Deposit date: | 2018-10-15 | | Release date: | 2019-10-30 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.29 Å) | | Cite: | The Two Deubiquitinating Enzymes fromChlamydia trachomatisHave Distinct Ubiquitin Recognition Properties.
Biochemistry, 59, 2020
|
|
3O4M
 
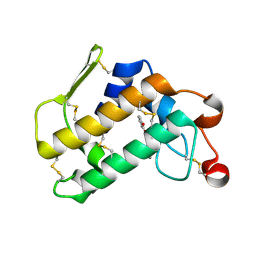 | | Crystal structure of porcine pancreatic phospholipase A2 in complex with 1,2-dihydroxybenzene | | Descriptor: | CALCIUM ION, CATECHOL, Phospholipase A2, ... | | Authors: | Dileep, K.V, Tintu, I, Karthe, P, Mandal, P.K, Haridas, M, Sadasivan, C. | | Deposit date: | 2010-07-27 | | Release date: | 2010-08-25 | | Last modified: | 2011-11-23 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Binding to PLA(2) may contribute to the anti-inflammatory activity of catechol
Chem.Biol.Drug Des., 2011
|
|
8DY8
 
 | |
4ETL
 
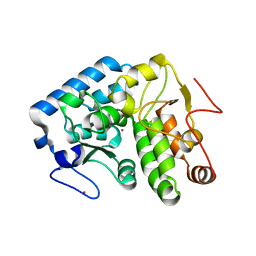 | | Crystallographic structure of phenylalanine hydroxylase from Chromobacterium violaceum F258A mutation | | Descriptor: | COBALT (II) ION, Phenylalanine-4-hydroxylase | | Authors: | Ronau, J.A, Paul, L.P, Corn, I.R, Wagner, K.T, Abu-Omar, M.M, Das, C. | | Deposit date: | 2012-04-24 | | Release date: | 2013-05-08 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.49 Å) | | Cite: | An additional substrate binding site in a bacterial phenylalanine hydroxylase.
Eur.Biophys.J., 42, 2013
|
|
4ESM
 
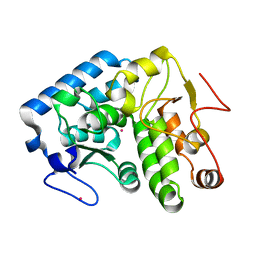 | | Crystallographic structure of phenylalanine hydroxylase from Chromobacterium violaceum Y155A mutation | | Descriptor: | COBALT (II) ION, Phenylalanine-4-hydroxylase | | Authors: | Ronau, J.A, Paul, L.P, Corn, I.R, Wagner, K.T, Abu-Omar, M.M, Das, C. | | Deposit date: | 2012-04-23 | | Release date: | 2013-05-08 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | An additional substrate binding site in a bacterial phenylalanine hydroxylase.
Eur.Biophys.J., 42, 2013
|
|
4DM9
 
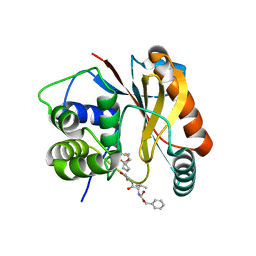 | | The Crystal Structure of Ubiquitin Carboxy-terminal hydrolase L1 (UCHL1) bound to a tripeptide fluoromethyl ketone Z-VAE(OMe)-FMK | | Descriptor: | Tripeptide fluoromethyl ketone inhibitor Z-VAE(OMe)-FMK, Ubiquitin carboxyl-terminal hydrolase isozyme L1 | | Authors: | Davies, C.W, Chaney, J, Korbel, G, Ringe, D, Petsko, G.A, Ploegh, H, Das, C. | | Deposit date: | 2012-02-07 | | Release date: | 2012-05-23 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | The co-crystal structure of ubiquitin carboxy-terminal hydrolase L1 (UCHL1) with a tripeptide fluoromethyl ketone (Z-VAE(OMe)-FMK).
Bioorg.Med.Chem.Lett., 22, 2012
|
|
5CRB
 
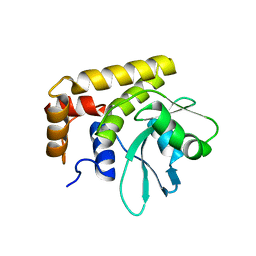 | | Crystal Structure of SdeA DUB | | Descriptor: | SdeA | | Authors: | Sheedlo, M.J, Qiu, J, Luo, Z.Q, Das, C. | | Deposit date: | 2015-07-22 | | Release date: | 2015-11-25 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural basis of substrate recognition by a bacterial deubiquitinase important for dynamics of phagosome ubiquitination.
Proc.Natl.Acad.Sci.USA, 112, 2015
|
|
5CRA
 
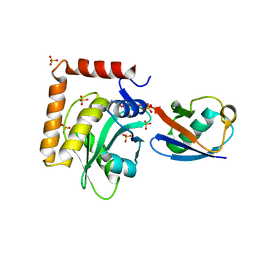 | | Structure of the SdeA DUB Domain | | Descriptor: | METHYL 4-AMINOBUTANOATE, Polyubiquitin-B, SULFATE ION, ... | | Authors: | Sheedlo, M.J, Qiu, J, Luo, Z.Q, Das, C. | | Deposit date: | 2015-07-22 | | Release date: | 2015-11-25 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.64 Å) | | Cite: | Structural basis of substrate recognition by a bacterial deubiquitinase important for dynamics of phagosome ubiquitination.
Proc.Natl.Acad.Sci.USA, 112, 2015
|
|
