7XUZ
 
 | | Crystal structure of a HDAC4-MEF2A-DNA ternary complex | | Descriptor: | DNA (5'-D(*TP*CP*TP*TP*AP*TP*AP*AP*AP*TP*AP*GP*T)-3'), DNA (5'-D(P*AP*CP*TP*AP*TP*TP*TP*AP*TP*AP*A)-3'), Histone deacetylase 4, ... | | Authors: | Dai, S.Y, Guo, L, Dey, R, Guo, M, Bates, D, Cayford, J, Chen, X.J, Wei, X.D, Chen, L, Chen, Y.H. | | Deposit date: | 2022-05-20 | | Release date: | 2023-11-29 | | Last modified: | 2024-06-12 | | Method: | X-RAY DIFFRACTION (3.591 Å) | | Cite: | Structural insights into the HDAC4-MEF2A-DNA complex and its implication in long-range transcriptional regulation.
Nucleic Acids Res., 52, 2024
|
|
7VNI
 
 | | AHR-ARNT PAS-B heterodimer | | Descriptor: | Ahr homolog spineless, Aryl hydrocarbon receptor nuclear translocator, SULFATE ION | | Authors: | Dai, S.Y. | | Deposit date: | 2021-10-11 | | Release date: | 2022-04-13 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.997 Å) | | Cite: | Structural insight into the ligand binding mechanism of aryl hydrocarbon receptor.
Nat Commun, 13, 2022
|
|
7VNA
 
 | | drosophlia AHR PAS-B domain | | Descriptor: | Ahr homolog spineless | | Authors: | Dai, S.Y. | | Deposit date: | 2021-10-10 | | Release date: | 2022-04-13 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.597 Å) | | Cite: | Structural insight into the ligand binding mechanism of aryl hydrocarbon receptor.
Nat Commun, 13, 2022
|
|
7VNH
 
 | |
7FJ2
 
 | | Structure of FOXM1 homodimer bound to a palindromic DNA site | | Descriptor: | DNA (5'-D(*AP*CP*CP*GP*TP*AP*AP*AP*CP*AP*TP*GP*TP*TP*TP*AP*CP*GP*GP*T)-3'), Forkhead box protein M1 | | Authors: | Dai, S.Y, Li, J, Zhang, H.J. | | Deposit date: | 2021-08-02 | | Release date: | 2022-01-26 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (3.098 Å) | | Cite: | Mechanistic Insights into the Preference for Tandem Binding Sites in DNA Recognition by FOXM1.
J.Mol.Biol., 434, 2021
|
|
7CBY
 
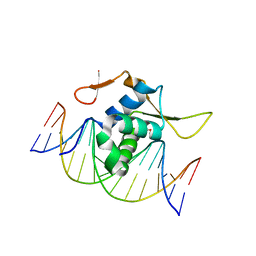 | | Structure of FOXG1 DNA binding domain bound to DBE2 DNA site | | Descriptor: | DI(HYDROXYETHYL)ETHER, DNA (5'-D(*CP*AP*AP*AP*AP*TP*GP*TP*AP*AP*AP*CP*AP*AP*GP*A)-3'), DNA (5'-D(*TP*CP*TP*TP*GP*TP*TP*TP*AP*CP*AP*TP*TP*TP*TP*G)-3'), ... | | Authors: | Dai, S.Y, Li, J, Chen, Y.H. | | Deposit date: | 2020-06-15 | | Release date: | 2020-10-28 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.646 Å) | | Cite: | Structural Basis for DNA Recognition by FOXG1 and the Characterization of Disease-causing FOXG1 Mutations.
J.Mol.Biol., 432, 2020
|
|
6LBM
 
 | |
6LBI
 
 | |
7V29
 
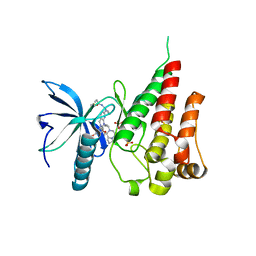 | | Crystal structure of FGFR4 with a dual-warhead covalent inhhibitor | | Descriptor: | Fibroblast growth factor receptor 4, N-[2-[[3-(3,5-dimethoxyphenyl)-2-oxidanylidene-1-[3-(4-propanoylpiperazin-1-yl)propyl]-4H-pyrimido[4,5-d]pyrimidin-7-yl]amino]phenyl]propanamide, SULFATE ION | | Authors: | Chen, X.J, Jiang, L.Y, Dai, S.Y, Qu, L.Z, Chen, Y.H. | | Deposit date: | 2021-08-07 | | Release date: | 2022-03-30 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.983 Å) | | Cite: | Structure-based design of a dual-warhead covalent inhibitor of FGFR4.
Commun Chem, 5, 2022
|
|
7WCT
 
 | | Crystal structure of FGFR4 kinase domain with 7v | | Descriptor: | Fibroblast growth factor receptor 4, GLYCEROL, SULFATE ION, ... | | Authors: | Chen, X.J, Lin, Q.M, Dai, S.Y, Chen, Y.H. | | Deposit date: | 2021-12-20 | | Release date: | 2022-03-30 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.106 Å) | | Cite: | Design, Synthesis, and Biological Evaluation of Aminoindazole Derivatives as Highly Selective Covalent Inhibitors of Wild-Type and Gatekeeper Mutant FGFR4.
J.Med.Chem., 65, 2022
|
|
7WCW
 
 | | Crystal structure of FGFR4(V550L) kinase domain with 7v | | Descriptor: | Fibroblast growth factor receptor 4, SULFATE ION, ~{N}-[2-[[5-[(1~{R})-1-[3,5-bis(chloranyl)pyridin-4-yl]ethoxy]-1~{H}-indazol-3-yl]amino]-3-fluoranyl-5-(4-morpholin-4-ylpiperidin-1-yl)phenyl]propanamide | | Authors: | Chen, X.J, Lin, Q.M, Dai, S.Y, Chen, Y.H. | | Deposit date: | 2021-12-20 | | Release date: | 2022-03-30 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.317 Å) | | Cite: | Design, Synthesis, and Biological Evaluation of Aminoindazole Derivatives as Highly Selective Covalent Inhibitors of Wild-Type and Gatekeeper Mutant FGFR4.
J.Med.Chem., 65, 2022
|
|
7WCX
 
 | | Crystal structure of FGFR4(V550M) kinase domain with 7v | | Descriptor: | Fibroblast growth factor receptor 4, SULFATE ION, ~{N}-[2-[[5-[(1~{R})-1-[3,5-bis(chloranyl)pyridin-4-yl]ethoxy]-1~{H}-indazol-3-yl]amino]-3-fluoranyl-5-(4-morpholin-4-ylpiperidin-1-yl)phenyl]propanamide | | Authors: | Chen, X.J, Lin, Q.M, Dai, S.Y, Chen, Y.H. | | Deposit date: | 2021-12-20 | | Release date: | 2022-03-30 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.175 Å) | | Cite: | Design, Synthesis, and Biological Evaluation of Aminoindazole Derivatives as Highly Selective Covalent Inhibitors of Wild-Type and Gatekeeper Mutant FGFR4.
J.Med.Chem., 65, 2022
|
|
7F3M
 
 | | Crystal structure of FGFR4 kinase domain with PRN1371 | | Descriptor: | 6-[2,6-bis(chloranyl)-3,5-dimethoxy-phenyl]-2-(methylamino)-8-[3-(4-prop-2-enoylpiperazin-1-yl)propyl]pyrido[2,3-d]pyrimidin-7-one, Fibroblast growth factor receptor 4, SULFATE ION | | Authors: | Chen, X.J, Qu, L.Z, Dai, S.Y, Wei, H.D, Chen, Y.H. | | Deposit date: | 2021-06-16 | | Release date: | 2022-01-12 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.289 Å) | | Cite: | Structural insights into the potency and selectivity of covalent pan-FGFR inhibitors
Commun Chem, 5, 2022
|
|
7DTZ
 
 | | FGFR4 complex with a covalent inhibitor | | Descriptor: | Fibroblast growth factor receptor 4, N-[2-[[5-[[2,6-bis(chloranyl)-3,5-dimethoxy-phenyl]methoxy]pyrimidin-2-yl]amino]-3-methyl-phenyl]-2-fluoranyl-prop-2-enamide, SULFATE ION | | Authors: | Chen, X.J, Dai, S.Y, Chen, Y.H. | | Deposit date: | 2021-01-07 | | Release date: | 2021-04-14 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.01 Å) | | Cite: | Investigation of Covalent Warheads in the Design of 2-Aminopyrimidine-based FGFR4 Inhibitors.
Acs Med.Chem.Lett., 12, 2021
|
|
