2APX
 
 | | Crystal Structure of the G17E/A52V/S54N/K66E/Q72H/E80V/L81S/T87S/G96V variant of the murine T cell receptor V beta 8.2 domain | | Descriptor: | MALONIC ACID, T cell receptor beta chain V | | Authors: | Cho, S, Swaminathan, C.P, Yang, J, Kerzic, M.C, Guan, R, Kieke, M.C, Kranz, D.M, Mariuzza, R.A, Sundberg, E.J. | | Deposit date: | 2005-08-16 | | Release date: | 2006-03-21 | | Last modified: | 2018-04-04 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural basis of affinity maturation and intramolecular cooperativity in a protein-protein interaction.
Structure, 13, 2005
|
|
2AW2
 
 | | Crystal structure of the human BTLA-HVEM complex | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, B and T lymphocyte attenuator, NICKEL (II) ION, ... | | Authors: | Compaan, D.M, Gonzalez, L.C, Tom, I, Loyet, K.M, Eaton, D, Hymowitz, S.G. | | Deposit date: | 2005-08-31 | | Release date: | 2005-09-27 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Attenuating Lymphocyte Activity: the crystal structure of the BTLA-HVEM complex
J.Biol.Chem., 280, 2005
|
|
2AQ2
 
 | | Crystal structure of T-cell receptor V beta domain variant complexed with superantigen SEC3 mutant | | Descriptor: | Enterotoxin type C-3, SODIUM ION, SULFATE ION, ... | | Authors: | Cho, S, Swaminathan, C.P, Yang, J, Kerzic, M.C, Guan, R, Kieke, M.C, Kranz, D.M, Mariuzza, R.A, Sundberg, E.J. | | Deposit date: | 2005-08-17 | | Release date: | 2006-03-21 | | Last modified: | 2017-10-11 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural basis of affinity maturation and intramolecular cooperativity in a protein-protein interaction.
Structure, 13, 2005
|
|
1TIE
 
 | |
2FJ9
 
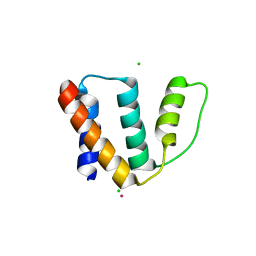 | | High resolution crystal structure of the unliganded human ACBP | | Descriptor: | Acyl-CoA-binding protein, CHLORIDE ION, LEAD (II) ION, ... | | Authors: | Taskinen, J.P, van Aalten, D.M, Knudsen, J, Wierenga, R.K. | | Deposit date: | 2006-01-02 | | Release date: | 2006-10-31 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | High resolution crystal structures of unliganded and liganded human liver ACBP reveal a new mode of binding for the acyl-CoA ligand.
Proteins, 66, 2006
|
|
2DAA
 
 | |
1N5Y
 
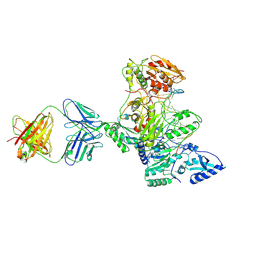 | | HIV-1 Reverse Transcriptase Crosslinked to Post-Translocation AZTMP-Terminated DNA (Complex P) | | Descriptor: | 5'-D(*A*CP*AP*GP*TP*CP*CP*CP*TP*GP*TP*TP*CP*GP*GP*(MRG)P*CP*GP*CP*CP*(ATM))-3', 5'-D(*AP*TP*GP*C*TP*AP*GP*GP*CP*GP*CP*CP*CP*GP*AP*AP*CP*AP*GP*GP*GP*AP*CP*TP*GP*TP*G)-3', MAGNESIUM ION, ... | | Authors: | Sarafianos, S.G, Clark Jr, A.D, Das, K, Tuske, S, Birktoft, J.J, Ilankumaran, P, Ramesha, A.R, Sayer, J.M, Jerina, D.M, Boyer, P.L, Hughes, S.H, Arnold, E. | | Deposit date: | 2002-11-07 | | Release date: | 2003-01-28 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Structure of HIV-1 Reverse Transcriptase with Pre-Translocation and Post-Translocation AZTMP-Terminated DNA
Embo J., 21, 2002
|
|
4BH7
 
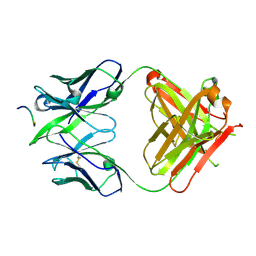 | |
4BH8
 
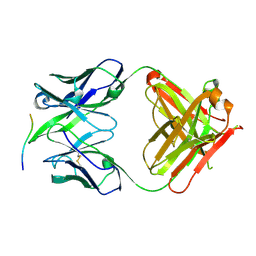 | |
4BKT
 
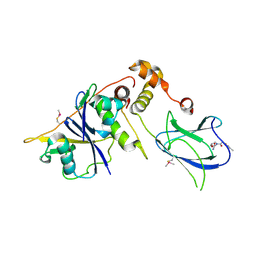 | | von Hippel Lindau protein:ElonginB:ElonginC complex, in complex with (2S,4R)-N-methyl-1-[2-(3-methyl-1,2-oxazol-5-yl)ethanoyl]-4-oxidanyl-pyrrolidine-2-carboxamide | | Descriptor: | (2S,4R)-N-methyl-1-[2-(3-methyl-1,2-oxazol-5-yl)ethanoyl]-4-oxidanyl-pyrrolidine-2-carboxamide, ARGININE, GLUTAMIC ACID, ... | | Authors: | Van Molle, I, Dias, D.M, Baud, M, Galdeano, C, Geraldes, C.F.G.C, Ciulli, A. | | Deposit date: | 2013-04-29 | | Release date: | 2013-11-27 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Is NMR Fragment Screening Fine-Tuned to Assess Druggability of Protein-Protein Interactions?
Acs Med.Chem.Lett., 5, 2014
|
|
1LW3
 
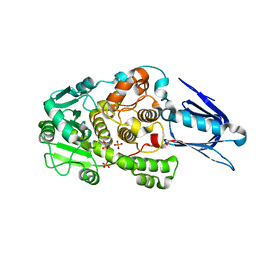 | | Crystal Structure of Myotubularin-related protein 2 complexed with phosphate | | Descriptor: | Myotubularin-related protein 2, PHOSPHATE ION | | Authors: | Begley, M.J, Taylor, G.S, Kim, S.-A, Veine, D.M, Dixon, J.E, Stuckey, J.A. | | Deposit date: | 2002-05-30 | | Release date: | 2003-10-07 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal Structure of a Phosphoinositide Phosphatase, MTMR2: Insights into Myotubular Myopathy and Charcot-Marie-Tooth Syndrome
Mol.Cell, 12, 2003
|
|
4C4N
 
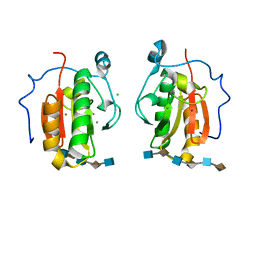 | | Crystal structure of the Sonic Hedgehog-heparin complex | | Descriptor: | 2-O-sulfo-alpha-L-idopyranuronic acid-(1-4)-2-deoxy-6-O-sulfo-2-(sulfoamino)-alpha-D-glucopyranose-(1-4)-2-O-sulfo-alpha-L-idopyranuronic acid-(1-4)-2-deoxy-6-O-sulfo-2-(sulfoamino)-alpha-D-glucopyranose-(1-4)-2-O-sulfo-alpha-L-idopyranuronic acid-(1-4)-2-deoxy-6-O-sulfo-2-(sulfoamino)-alpha-D-glucopyranose, 2-deoxy-6-O-sulfo-2-(sulfoamino)-alpha-D-glucopyranose-(1-4)-2-O-sulfo-alpha-L-idopyranuronic acid-(1-4)-2-deoxy-6-O-sulfo-2-(sulfoamino)-alpha-D-glucopyranose-(1-4)-2-O-sulfo-alpha-L-idopyranuronic acid-(1-4)-2-deoxy-6-O-sulfo-2-(sulfoamino)-alpha-D-glucopyranose-(1-4)-2-O-sulfo-alpha-L-idopyranuronic acid, CALCIUM ION, ... | | Authors: | Whalen, D.M, Malinauskas, T, Gilbert, R.J.C, Siebold, C. | | Deposit date: | 2013-09-05 | | Release date: | 2013-10-02 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.36 Å) | | Cite: | Structural Insights Into Proteoglycan-Shaped Hedgehog Signaling.
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
4BQF
 
 | | Arabidopsis thaliana cytosolic alpha-1,4-glucan phosphorylase (PHS2) in complex with acarbose | | Descriptor: | 4,6-dideoxy-4-{[(1S,4R,5S,6S)-4,5,6-trihydroxy-3-(hydroxymethyl)cyclohex-2-en-1-yl]amino}-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose-(1-4)-beta-D-glucopyranose, ALPHA-GLUCAN PHOSPHORYLASE 2, CYTOSOLIC, ... | | Authors: | O'Neill, E.C, Rashid, A.M, Stevenson, C.E.M, Hetru, A.C, Gunning, A.P, Rejzek, M, Nepogodiev, S.A, Bornemann, S, Lawson, D.M, Field, R.A. | | Deposit date: | 2013-05-30 | | Release date: | 2014-02-19 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Sugar-Coated Sensor Chip and Nanoparticle Surfaces for the in Vitro Enzymatic Synthesis of Starch-Like Materials
Chem.Sci., 5, 2014
|
|
3L29
 
 | | Crystal Structure of Zaire Ebola VP35 interferon inhibitory domain K319A/R322A mutant | | Descriptor: | CHLORIDE ION, Polymerase cofactor VP35 | | Authors: | Leung, D.W, Ramanan, P, Borek, D.M, Amarasinghe, G.K. | | Deposit date: | 2009-12-14 | | Release date: | 2010-02-02 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Mutations abrogating VP35 interaction with double-stranded RNA render ebola virus avirulent in guinea pigs.
J.Virol., 84, 2010
|
|
3GJL
 
 | | crystal structure of a DNA duplex containing 7,8-dihydropyridol[2,3-d]pyrimidin-2-one | | Descriptor: | 5'-D(*CP*GP*CP*GP*AP*A)-3', 5'-D(P*TP*TP*(B7C)P*GP*CP*G)-3', SODIUM ION | | Authors: | Takenaka, A, Juan, E.C.M, Shimizu, S, Haraguchi, T, Xiao, M, Kurose, T, Ohkubo, A, Sekine, M, Shibata, T, Millington, C.L, Williams, D.M. | | Deposit date: | 2009-03-09 | | Release date: | 2010-03-31 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.92 Å) | | Cite: | Insights into the stabilizing contributions of bicyclic cytosine analogues: crystal structures of DNA duplexes containing 7,8-dihydropyridol[2,3-d]pyrimidin-2-one
To be Published
|
|
4CDL
 
 | | Crystal Structure of Retro-aldolase RA110.4-6 Complexed with Inhibitor 1-(6-methoxy-2-naphthalenyl)-1,3-butanedione | | Descriptor: | (2E)-1-(6-methoxynaphthalen-2-yl)but-2-en-1-one, STEROID DELTA-ISOMERASE | | Authors: | Pinkas, D.M, Studer, S, Obexer, R, Giger, L, Gruetter, M.G, Baker, D, Hilvert, D. | | Deposit date: | 2013-11-01 | | Release date: | 2014-11-12 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Active Site Plasticity of a Computationally Designed Retro-Aldolase Enzyme
To be Published
|
|
4BQI
 
 | | ARABIDOPSIS THALIANA cytosolic alpha-1,4-glucan phosphorylase (PHS2) in complex with maltotriose | | Descriptor: | ALPHA-GLUCAN PHOSPHORYLASE 2, CYTOSOLIC, DI(HYDROXYETHYL)ETHER, ... | | Authors: | O'Neill, E.C, Rashid, A.M, Stevenson, C.E.M, Hetru, A.C, Gunning, A.P, Rejzek, M, Nepogodiev, S.A, Bornemann, S, Lawson, D.M, Field, R.A. | | Deposit date: | 2013-05-30 | | Release date: | 2014-02-19 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Sugar-Coated Sensor Chip and Nanoparticle Surfaces for the in Vitro Enzymatic Synthesis of Starch-Like Materials
Chem.Sci., 5, 2014
|
|
4C4M
 
 | | Crystal structure of the Sonic Hedgehog-chondroitin-4-sulphate complex | | Descriptor: | ACETATE ION, CALCIUM ION, SONIC HEDGEHOG PROTEIN, ... | | Authors: | Whalen, D.M, Malinauskas, T, Gilbert, R.J.C, Siebold, C. | | Deposit date: | 2013-09-05 | | Release date: | 2013-10-02 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.74 Å) | | Cite: | Structural Insights Into Proteoglycan-Shaped Hedgehog Signaling.
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
4CAU
 
 | | THREE-DIMENSIONAL STRUCTURE OF DENGUE VIRUS SEROTYPE 1 COMPLEXED WITH 2 HMAB 14C10 FAB | | Descriptor: | ENVELOPE PROTEIN E, FAB 14C10 | | Authors: | Teoh, E.P, Kukkaro, P, Teo, E.W, Lim, A.P, Tan, T.T, Yip, A, Schul, W, Aung, M, Kostyuchenko, V.A, Leo, Y.S, Chan, S.H, Smith, K.G, Chan, A.H, Zou, G, Ooi, E.E, Kemeny, D.M, Tan, G.K, Ng, J.K, Ng, M.L, Alonso, S, Fisher, D, Shi, P.Y, Hanson, B.J, Lok, S.M, Macary, P.A. | | Deposit date: | 2013-10-09 | | Release date: | 2013-10-16 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (7 Å) | | Cite: | The Structural Basis for Serotype-Specific Neutralization of Dengue Virus by a Human Antibody.
Sci.Trans.Med, 4, 2012
|
|
1N6Q
 
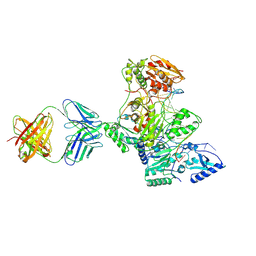 | | HIV-1 Reverse Transcriptase Crosslinked to pre-translocation AZTMP-terminated DNA (complex N) | | Descriptor: | 5'-D(*A*CP*AP*GP*TP*CP*CP*CP*TP*GP*TP*TP*CP*GP*GP*(MRG)P*CP*GP*CP*CP*AP*(ATM))-3', 5'-D(*AP*T*GP*CP*AP*TP*GP*GP*CP*GP*CP*CP*CP*GP*AP*AP*CP*AP*GP*GP*GP*AP*CP*TP*GP*TP*G)-3', MAGNESIUM ION, ... | | Authors: | Sarafianos, S.G, Clark Jr, A.D, Das, K, Tuske, S, Birktoft, J.J, Ilankumaran, I, Ramesha, A.R, Sayer, J.M, Jerina, D.M, Boyer, P.L, Hughes, S.H, Arnold, E. | | Deposit date: | 2002-11-11 | | Release date: | 2003-01-14 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structures of HIV-1 Reverse Transcriptase with Pre- and Post-translocation AZTMP-terminated DNA
Embo J., 21, 2002
|
|
1NWA
 
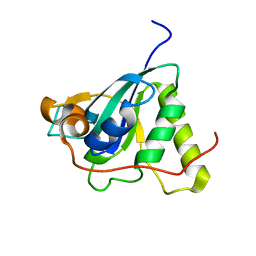 | | Structure of Mycobacterium tuberculosis Methionine Sulfoxide Reductase A in Complex with Protein-bound Methionine | | Descriptor: | Peptide methionine sulfoxide reductase msrA | | Authors: | Taylor, A.B, Benglis Jr, D.M, Dhandayuthapani, S, Hart, P.J, TB Structural Genomics Consortium (TBSGC) | | Deposit date: | 2003-02-05 | | Release date: | 2003-07-08 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structure of Mycobacterium tuberculosis Methionine Sulfoxide Reductase A in Complex with Protein-bound Methionine
J.Bacteriol., 185, 2003
|
|
4C7A
 
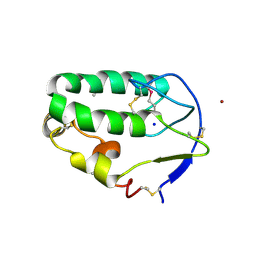 | | Crystal structure of the Smoothened CRD, selenomethionine-labeled | | Descriptor: | SMOOTHENED, SODIUM ION, ZINC ION | | Authors: | Nachtergaele, S, Whalen, D.M, Mydock, L.K, Zhao, Z, Malinauskas, T, Krishnan, K, Ingham, P.W, Covey, D.F, Rohatgi, R, Siebold, C. | | Deposit date: | 2013-09-20 | | Release date: | 2013-11-06 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structure and Function of the Smoothened Extracellular Domain in Vertebrate Hedgehog Signaling
Elife, 2, 2013
|
|
1OD0
 
 | | Family 1 b-glucosidase from Thermotoga maritima | | Descriptor: | BETA-GLUCOSIDASE A | | Authors: | Gloster, T, Zechel, D.L, Boraston, A.B, Boraston, C.M, Macdonald, J.M, Tilbrook, D.M, Stick, R.V, Davies, G.J. | | Deposit date: | 2003-02-12 | | Release date: | 2003-03-06 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.11 Å) | | Cite: | Iminosugar Glycosidase Inhibitors: Structural and Thermodynamic Dissection of the Binding of Isofagomine and 1-Deoxynojirimycin to Beta-Glucosidases
J.Am.Chem.Soc., 125, 2003
|
|
4CAG
 
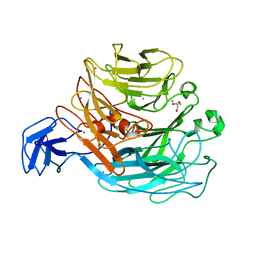 | | Bacillus licheniformis Rhamnogalacturonan Lyase PL11 | | Descriptor: | CALCIUM ION, GLYCEROL, POLYSACCHARIDE LYASE FAMILY 11 PROTEIN | | Authors: | Otten, H, Rodrigues da Silva, I.I.C, Jers, C, Nyffenegger, C, Larsen, D.M, Mikkelsen, J.D, Larsen, S. | | Deposit date: | 2013-10-08 | | Release date: | 2014-10-15 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.498 Å) | | Cite: | Design of Thermostable Rhamnogalacturonan Lyase Mutants from Bacillus Licheniformis by Combination of Targeted Single Point Mutations.
Appl.Microbiol.Biotechnol., 98, 2014
|
|
4C79
 
 | | Crystal structure of the Smoothened CRD, native | | Descriptor: | SMOOTHENED, SODIUM ION, ZINC ION | | Authors: | Nachtergaele, S, Whalen, D.M, Mydock, L.K, Zhao, Z, Malinauskas, T, Krishnan, K, Ingham, P.W, Covey, D.F, Rohatgi, R, Siebold, C. | | Deposit date: | 2013-09-20 | | Release date: | 2013-11-06 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.604 Å) | | Cite: | Structure and Function of the Smoothened Extracellular Domain in Vertebrate Hedgehog Signaling
Elife, 2, 2013
|
|
