8E75
 
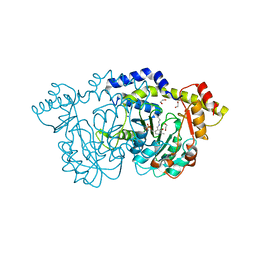 | | Crystal structure of Pcryo_0616, the aminotransferase required to synthesize UDP-N-acetyl-3-amino-D-glucosaminuronic acid (UDP-GlcNAc3NA) | | Descriptor: | 1,2-ETHANEDIOL, DegT/DnrJ/EryC1/StrS aminotransferase, SODIUM ION | | Authors: | Hofmeister, D.L, Seltzner, C.A, Bockhaus, N.J, thoden, J.B, Holden, H.M. | | Deposit date: | 2022-08-23 | | Release date: | 2022-11-23 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.25 Å) | | Cite: | Investigation of the enzymes required for the biosynthesis of 2,3-diacetamido-2,3-dideoxy-d-glucuronic acid in Psychrobacter cryohalolentis K5 T.
Protein Sci., 32, 2023
|
|
8E62
 
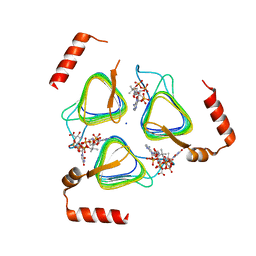 | | STRUCTURE OF Pcryo_0615 from Psychrobacter cryohalolentis, an N-acetyltransferase required to produce Diacetamido-2,3-dideoxy-D-glucuronic acid | | Descriptor: | (2S,3S,4R,5R,6R)-5-(acetylamino)-4-amino-6-{[(R)-{[(R)-{[(2R,3S,4R,5R)-5-(2,4-dioxo-3,4-dihydropyrimidin-1(2H)-yl)-3,4-dihydroxytetrahydrofuran-2-yl]methoxy}(hydroxy)phosphoryl]oxy}(hydroxy)phosphoryl]oxy}-3-hydroxytetrahydro-2H-pyran-2-carboxylic acid, COENZYME A, SODIUM ION, ... | | Authors: | Hofmeister, D.L, Bockhaus, N.J, Seltzner, C.A, Thoden, J.B, Holden, H.M. | | Deposit date: | 2022-08-22 | | Release date: | 2022-11-23 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Investigation of the enzymes required for the biosynthesis of 2,3-diacetamido-2,3-dideoxy-d-glucuronic acid in Psychrobacter cryohalolentis K5 T.
Protein Sci., 32, 2023
|
|
5K6K
 
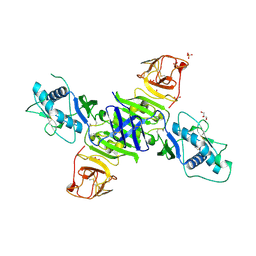 | | Zika virus non-structural protein 1 (NS1) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, GLYCEROL, SULFATE ION, ... | | Authors: | Akey, D.L, Brown, W.C, Smith, J.L. | | Deposit date: | 2016-05-24 | | Release date: | 2016-07-06 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.89 Å) | | Cite: | Extended surface for membrane association in Zika virus NS1 structure.
Nat.Struct.Mol.Biol., 23, 2016
|
|
5KN7
 
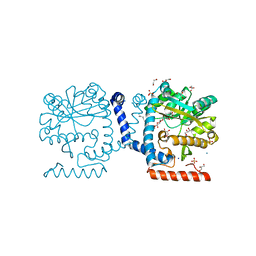 | | Lipid A secondary acyltransferase LpxM from Acinetobacter baumannii | | Descriptor: | DODECYL-BETA-D-MALTOSIDE, GLYCEROL, Lipid A biosynthesis lauroyl acyltransferase, ... | | Authors: | Dovala, D.L, Hu, Q, Metzger IV, L.E. | | Deposit date: | 2016-06-27 | | Release date: | 2016-09-28 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.99 Å) | | Cite: | Structure-guided enzymology of the lipid A acyltransferase LpxM reveals a dual activity mechanism.
Proc.Natl.Acad.Sci.USA, 113, 2016
|
|
5KNK
 
 | | Lipid A secondary acyltransferase LpxM from Acinetobacter baumannii with catalytic residue substitution (E127A) | | Descriptor: | DODECYL-BETA-D-MALTOSIDE, GLYCEROL, Lipid A biosynthesis lauroyl acyltransferase, ... | | Authors: | Dovala, D.L, Hu, Q, Metzger IV, L.E. | | Deposit date: | 2016-06-28 | | Release date: | 2016-09-28 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structure-guided enzymology of the lipid A acyltransferase LpxM reveals a dual activity mechanism.
Proc.Natl.Acad.Sci.USA, 113, 2016
|
|
5KZL
 
 | | Structure of Heme Oxygenase from Leptospira interrogans | | Descriptor: | Heme oxygenase, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Klinke, S, Soldano, A, Otero, L.H, Rivera, M, Catalano-Dupuy, D.L, Ceccarelli, E.A. | | Deposit date: | 2016-07-25 | | Release date: | 2017-07-26 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.73 Å) | | Cite: | Structural and mutational analyses of the Leptospira interrogans virulence-related heme oxygenase provide insights into its catalytic mechanism.
PLoS ONE, 12, 2017
|
|
1KJU
 
 | | Ca2+-ATPase in the E2 State | | Descriptor: | Sarcoplasmic/endoplasmic reticulum calcium ATPase 1a | | Authors: | Xu, C, Rice, W.J, He, W, Stokes, D.L. | | Deposit date: | 2001-12-05 | | Release date: | 2001-12-19 | | Last modified: | 2024-02-14 | | Method: | ELECTRON MICROSCOPY (6 Å) | | Cite: | A structural model for the catalytic cycle of Ca(2+)-ATPase.
J.Mol.Biol., 316, 2002
|
|
1EFP
 
 | | ELECTRON TRANSFER FLAVOPROTEIN (ETF) FROM PARACOCCUS DENITRIFICANS | | Descriptor: | ADENOSINE MONOPHOSPHATE, FLAVIN-ADENINE DINUCLEOTIDE, PROTEIN (ELECTRON TRANSFER FLAVOPROTEIN) | | Authors: | Roberts, D.L, Salazar, D, Fulmer, J.P, Frerman, F.E, Kim, J.J.-P. | | Deposit date: | 1998-12-18 | | Release date: | 1999-08-09 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Crystal structure of Paracoccus denitrificans electron transfer flavoprotein: structural and electrostatic analysis of a conserved flavin binding domain.
Biochemistry, 38, 1999
|
|
6JD1
 
 | | Cryo-EM Structure of Sulfolobus solfataricus ketol-acid reductoisomerase (Sso-KARI) in complex with Mg2+, NADH, and CPD at pH7.5 | | Descriptor: | 1,4-DIHYDRONICOTINAMIDE ADENINE DINUCLEOTIDE, MAGNESIUM ION, Putative ketol-acid reductoisomerase 2, ... | | Authors: | Chen, C.Y, Chang, Y.C, Lin, K.F, Huang, C.H, Lin, B.L, Ko, T.P, Hsieh, D.L, Tsai, M.D. | | Deposit date: | 2019-01-30 | | Release date: | 2019-04-17 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (3.38 Å) | | Cite: | Use of Cryo-EM To Uncover Structural Bases of pH Effect and Cofactor Bispecificity of Ketol-Acid Reductoisomerase.
J. Am. Chem. Soc., 141, 2019
|
|
6RJ0
 
 | |
6JD2
 
 | | Crystal structure of Sulfolobus solfataricus ketol-acid reductoisomerase (Sso-KARI) in complex with Mg2+ at pH8.5 | | Descriptor: | BETA-MERCAPTOETHANOL, MAGNESIUM ION, Putative ketol-acid reductoisomerase 2 | | Authors: | Chen, C.Y, Chang, Y.C, Lin, K.F, Huang, C.H, Lin, B.L, Ko, T.P, Hsieh, D.L, Tsai, M.D. | | Deposit date: | 2019-01-30 | | Release date: | 2019-08-14 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.53 Å) | | Cite: | Use of Cryo-EM To Uncover Structural Bases of pH Effect and Cofactor Bispecificity of Ketol-Acid Reductoisomerase.
J.Am.Chem.Soc., 141, 2019
|
|
6JCW
 
 | | Cryo-EM Structure of Sulfolobus solfataricus ketol-acid reductoisomerase (Sso-KARI) with Mg2+ at pH8.5 | | Descriptor: | MAGNESIUM ION, ketol-acid reductoisomerase | | Authors: | Chen, C.Y, Chang, Y.C, Lin, K.F, Huang, C.H, Lin, B.L, Ko, T.P, Hsieh, D.L, Tsai, M.D. | | Deposit date: | 2019-01-30 | | Release date: | 2019-04-17 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (3.04 Å) | | Cite: | Use of Cryo-EM To Uncover Structural Bases of pH Effect and Cofactor Bispecificity of Ketol-Acid Reductoisomerase.
J. Am. Chem. Soc., 141, 2019
|
|
6RTJ
 
 | | Thioredoxin glutathione reductase from Schistosoma mansoni in complex with 1-[(dimethylamino)methyl]-2-naphthol at 1 hour of soaking | | Descriptor: | 1-[(dimethylamino)methyl]naphthalen-2-ol, CALCIUM ION, FLAVIN-ADENINE DINUCLEOTIDE, ... | | Authors: | Angelucci, F, Silvestri, I, Fata, F, Williams, D.L. | | Deposit date: | 2019-05-24 | | Release date: | 2020-01-08 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Ectopic suicide inhibition of thioredoxin glutathione reductase.
Free Radic Biol Med, 147, 2020
|
|
6RTO
 
 | | Thioredoxin glutathione reductase from Schistosoma mansoni in complex with 1-[(dimethylamino)methyl]-2-naphthol at 4 hours of soaking | | Descriptor: | 1-methylidenenaphthalen-2-one, FLAVIN-ADENINE DINUCLEOTIDE, TRIETHYLENE GLYCOL, ... | | Authors: | Angelucci, F, Silvestri, I, Fata, F, Williams, D.L. | | Deposit date: | 2019-05-24 | | Release date: | 2020-01-08 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Ectopic suicide inhibition of thioredoxin glutathione reductase.
Free Radic Biol Med, 147, 2020
|
|
6JCZ
 
 | | Cryo-EM Structure of Sulfolobus solfataricus ketol-acid reductoisomerase (Sso-KARI) in complex with Mg2+, NADPH, and CPD at pH7.5 | | Descriptor: | MAGNESIUM ION, NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, Putative ketol-acid reductoisomerase 2, ... | | Authors: | Chen, C.Y, Chang, Y.C, Lin, K.F, Huang, C.H, Lin, B.L, Ko, T.P, Hsieh, D.L, Tsai, M.D. | | Deposit date: | 2019-01-30 | | Release date: | 2019-04-17 | | Last modified: | 2019-05-01 | | Method: | ELECTRON MICROSCOPY (3.35 Å) | | Cite: | Use of Cryo-EM To Uncover Structural Bases of pH Effect and Cofactor Bispecificity of Ketol-Acid Reductoisomerase.
J. Am. Chem. Soc., 141, 2019
|
|
6RTM
 
 | | Thioredoxin glutathione reductase from Schistosoma mansoni in complex with 1-[(dimethylamino)methyl]-2-naphthol at 2 hour of soaking | | Descriptor: | 1-methylidenenaphthalen-2-one, FLAVIN-ADENINE DINUCLEOTIDE, POTASSIUM ION, ... | | Authors: | Angelucci, F, Silvestri, I, Fata, F, Williams, D.L. | | Deposit date: | 2019-05-24 | | Release date: | 2020-01-08 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Ectopic suicide inhibition of thioredoxin glutathione reductase.
Free Radic Biol Med, 147, 2020
|
|
6S6L
 
 | | Cryo-EM structure of murine norovirus (MNV-1) | | Descriptor: | Capsid protein | | Authors: | Snowden, J.S, Hurdiss, D.L, Adeyemi, O.O, Ranson, N.A, Herod, M.R, Stonehouse, N.J. | | Deposit date: | 2019-07-03 | | Release date: | 2020-05-13 | | Last modified: | 2024-05-22 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Dynamics in the murine norovirus capsid revealed by high-resolution cryo-EM.
Plos Biol., 18, 2020
|
|
6JCV
 
 | | Cryo-EM structure of Sulfolobus solfataricus ketol-acid reductoisomerase (Sso-KARI) with Mg2+ at pH7.5 | | Descriptor: | MAGNESIUM ION, Putative ketol-acid reductoisomerase 2 | | Authors: | Chen, C.Y, Chang, Y.C, Lin, K.F, Huang, C.H, Lin, B.L, Ko, T.P, Hsieh, D.L, Tsai, M.D. | | Deposit date: | 2019-01-30 | | Release date: | 2019-04-17 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (2.92 Å) | | Cite: | Use of Cryo-EM To Uncover Structural Bases of pH Effect and Cofactor Bispecificity of Ketol-Acid Reductoisomerase.
J. Am. Chem. Soc., 141, 2019
|
|
1QHM
 
 | | ESCHERICHIA COLI PYRUVATE FORMATE LYASE LARGE DOMAIN | | Descriptor: | PYRUVATE FORMATE-LYASE | | Authors: | Leppanen, V.-M, Merckel, M.C, Ollis, D.L, Wong, K.K, Kozarich, J.W, Goldman, A. | | Deposit date: | 1999-05-19 | | Release date: | 2000-05-24 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Pyruvate formate lyase is structurally homologous to type I ribonucleotide reductase.
Structure Fold.Des., 7, 1999
|
|
1QN1
 
 | | SOLUTION STRUCTURE OF DESULFOVIBRIO GIGAS FERRICYTOCHROME C3, NMR, 15 STRUCTURES | | Descriptor: | CYTOCHROME C3, HEME C | | Authors: | Brennan, L, Messias, A.C, Legall, J, Turner, D.L, Xavier, A.V. | | Deposit date: | 1999-10-11 | | Release date: | 2000-10-12 | | Last modified: | 2024-10-09 | | Method: | SOLUTION NMR | | Cite: | Structural Basis for the Network of Functional Cooperativities in Cytochromes C3 from Desulfovibrio Gigas: Solution Structures of the Oxidised and Reduced States
J.Mol.Biol., 298, 2000
|
|
4M65
 
 | | In situ thermolysin crystallized on a MiTeGen micromesh with asparagine ligand | | Descriptor: | 1,2-ETHANEDIOL, ASPARAGINE, CALCIUM ION, ... | | Authors: | Yin, X, Scalia, A, Leroy, L, Cuttitta, C.M, Polizzo, G.M, Ericson, D.L, Roessler, C.G, Campos, O, Agarwal, R, Allaire, M, Orville, A.M, Jackimowicz, R, Ma, M.Y, Sweet, R.M, Soares, A.S. | | Deposit date: | 2013-08-08 | | Release date: | 2013-10-30 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Hitting the target: fragment screening with acoustic in situ
co-crystallization of proteins plus fragment libraries on
pin-mounted data-collection micromeshes
Acta Crystallogr.,Sect.D, D70
|
|
4LTQ
 
 | | Bacterial sodium channel in low calcium, P42 space group | | Descriptor: | Ion transport protein | | Authors: | Shaya, D, Findeisen, F, Abderemane-Ali, F, Arrigoni, C, Wong, S, Reddy Nurva, S, Loussouarn, G, Minor, D.L. | | Deposit date: | 2013-07-23 | | Release date: | 2013-10-23 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (5.5 Å) | | Cite: | Structure of a prokaryotic sodium channel pore reveals essential gating elements and an outer ion binding site common to eukaryotic channels.
J.Mol.Biol., 426, 2014
|
|
6PBO
 
 | | Staphylococcus aureus Dihydrofolate reductase in complex with NADPH and UCP1232 | | Descriptor: | (4-{6-[(2S)-4-(2,4-diamino-6-ethylpyrimidin-5-yl)but-3-yn-2-yl]-2H-1,3-benzodioxol-4-yl}phenyl)acetic acid, Dihydrofolate reductase, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, ... | | Authors: | Reeve, S.M, Wright, D.L. | | Deposit date: | 2019-06-14 | | Release date: | 2019-10-23 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.649 Å) | | Cite: | Toward Broad Spectrum Dihydrofolate Reductase Inhibitors Targeting Trimethoprim Resistant Enzymes Identified in Clinical Isolates of Methicillin ResistantStaphylococcus aureus.
Acs Infect Dis., 5, 2019
|
|
4LWS
 
 | | EsxA : EsxB (SeMet) hetero-dimer from Thermomonospora curvata | | Descriptor: | ACETATE ION, GLYCEROL, Uncharacterized protein | | Authors: | Dovala, D.L, Cox, J.S, Stroud, R.M, Rosenberg, O.S. | | Deposit date: | 2013-07-28 | | Release date: | 2015-02-04 | | Last modified: | 2016-09-21 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Substrates Control Multimerization and Activation of the Multi-Domain ATPase Motor of Type VII Secretion.
Cell(Cambridge,Mass.), 161, 2015
|
|
1QBH
 
 | | SOLUTION STRUCTURE OF A BACULOVIRAL INHIBITOR OF APOPTOSIS (IAP) REPEAT | | Descriptor: | INHIBITOR OF APOPTOSIS PROTEIN (2MIHB/C-IAP-1), ZINC ION | | Authors: | Hinds, M.G, Norton, R.S, Vaux, D.L, Day, C.L. | | Deposit date: | 1999-04-20 | | Release date: | 1999-10-20 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution structure of a baculoviral inhibitor of apoptosis (IAP) repeat.
Nat.Struct.Biol., 6, 1999
|
|
