4U3G
 
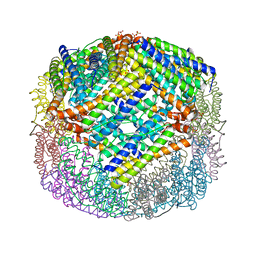 | | Crystal structure of Escherichia coli bacterioferritin mutant D132F | | Descriptor: | Bacterioferritin, SULFATE ION | | Authors: | Wong, S.G, Grigg, J.C, Le Brun, N.E, Moore, G.R, Murphy, M.E.P, Mauk, A.G. | | Deposit date: | 2014-07-21 | | Release date: | 2014-12-24 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The B-type Channel Is a Major Route for Iron Entry into the Ferroxidase Center and Central Cavity of Bacterioferritin.
J.Biol.Chem., 290, 2015
|
|
5JH5
 
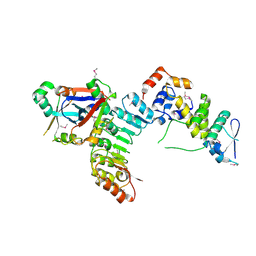 | | Structural Basis for the Hierarchical Assembly of the Core of PRC1.1 | | Descriptor: | BCL-6 corepressor-like protein 1, Lysine-specific demethylase 2B, Polycomb group RING finger protein 1, ... | | Authors: | Wong, S.J, Taylor, A.B, Hart, P.J, Kim, C.A. | | Deposit date: | 2016-04-20 | | Release date: | 2016-09-14 | | Last modified: | 2016-10-19 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | KDM2B Recruitment of the Polycomb Group Complex, PRC1.1, Requires Cooperation between PCGF1 and BCORL1.
Structure, 24, 2016
|
|
4U4J
 
 | |
4U59
 
 | |
4U48
 
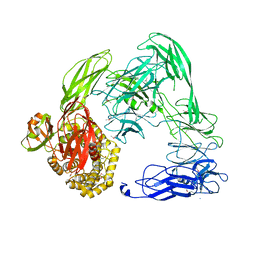 | |
3E2C
 
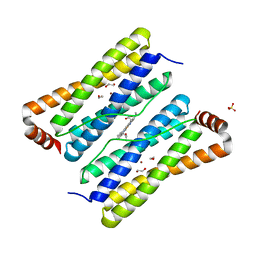 | | Escherichia coli Bacterioferritin Mutant E128R/E135R | | Descriptor: | 1,2-ETHANEDIOL, Bacterioferritin, PROTOPORPHYRIN IX CONTAINING FE, ... | | Authors: | Wong, S.G, Tom-Yew, S.A.L, Lewin, A, Le Brun, N.E, Moore, G.R, Murphy, M.E.P, Mauk, A.G. | | Deposit date: | 2008-08-05 | | Release date: | 2009-05-12 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural and Mechanistic Studies of a Stabilized Subunit Dimer Variant of Escherichia coli Bacterioferritin Identify Residues Required for Core Formation.
J.Biol.Chem., 284, 2009
|
|
5EOD
 
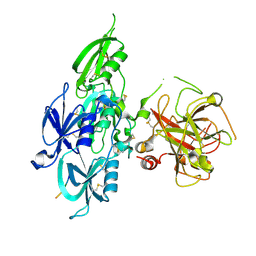 | | Human Plasma Coagulation FXI with peptide LP2 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Coagulation factor XI, LP2 | | Authors: | Wong, S.S, Ostergaard, S, Hall, G, Li, C, Williams, P.M, Stennicke, H, Emsley, J. | | Deposit date: | 2015-11-10 | | Release date: | 2016-04-06 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | A novel DFP tripeptide motif interacts with the coagulation factor XI apple 2 domain.
Blood, 127, 2016
|
|
5EOK
 
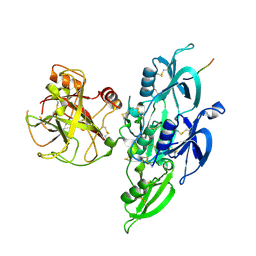 | | Human Plasma Coagulation Factor XI in complex with peptide P39 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, Coagulation factor XI, ... | | Authors: | Wong, S.S, Ostergaard, S, Hall, G, Li, C, Williams, P.M, Stennicke, H, Emsley, J. | | Deposit date: | 2015-11-10 | | Release date: | 2016-04-06 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | A novel DFP tripeptide motif interacts with the coagulation factor XI apple 2 domain.
Blood, 127, 2016
|
|
3ULK
 
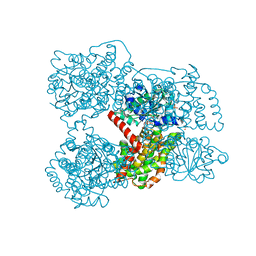 | | E. coli Ketol-acid reductoisomerase in complex with NADPH and Mg2+ | | Descriptor: | Ketol-acid reductoisomerase, MAGNESIUM ION, NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, ... | | Authors: | Wong, S.H, Lonhienne, T.G.A, Winzor, D.J, Schenk, G, Guddat, L.W. | | Deposit date: | 2011-11-11 | | Release date: | 2012-10-17 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Bacterial and plant ketol-acid reductoisomerases have different mechanisms of induced fit during the catalytic cycle
J.Mol.Biol., 424, 2012
|
|
2N1L
 
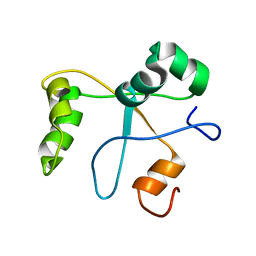 | | Solution structure of the BCOR PUFD | | Descriptor: | BCL-6 corepressor | | Authors: | Wong, S.J, Gearhart, M.D, Ha, D.J, Corcoran, C.M, Diaz, V, Taylor, A.B, Schirf, V, Ilangovan, U, Hinck, A.P, Demeler, B, Hart, J, Bardwell, V.J, Kim, C.A. | | Deposit date: | 2015-04-06 | | Release date: | 2016-04-06 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | Structural basis for the hierarchical assembly of the core of PRC1.1
To be Published
|
|
7SA8
 
 | |
5I25
 
 | |
6VJK
 
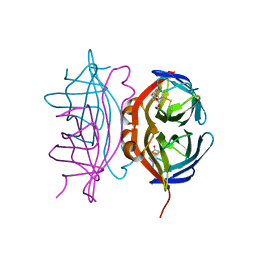 | | Streptavidin mutant M88 (N49C/A86C) | | Descriptor: | BIOTIN, Streptavidin | | Authors: | Marangoni, J.M, Wu, S.C, Fogen, D, Wong, S.L, Ng, K.K.S. | | Deposit date: | 2020-01-16 | | Release date: | 2020-12-23 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Engineering a disulfide-gated switch in streptavidin enables reversible binding without sacrificing binding affinity.
Sci Rep, 10, 2020
|
|
6VJL
 
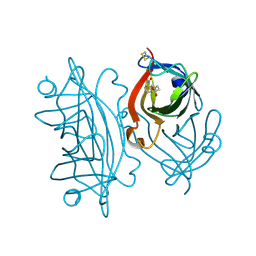 | | Streptavidin mutant M112 (G26C/A46C) | | Descriptor: | BIOTIN, Streptavidin | | Authors: | Marangoni, J.M, Wu, S.C, Fogen, D, Wong, S.L, Ng, K.K.S. | | Deposit date: | 2020-01-16 | | Release date: | 2020-12-23 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Engineering a disulfide-gated switch in streptavidin enables reversible binding without sacrificing binding affinity.
Sci Rep, 10, 2020
|
|
1LXL
 
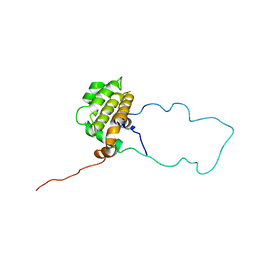 | | NMR STRUCTURE OF BCL-XL, AN INHIBITOR OF PROGRAMMED CELL DEATH, MINIMIZED AVERAGE STRUCTURE | | Descriptor: | BCL-XL | | Authors: | Muchmore, S.W, Sattler, M, Liang, H, Meadows, R.P, Harlan, J.E, Yoon, H.S, Nettesheim, D, Chang, B.S, Thompson, C.B, Wong, S.L, Ng, S.C, Fesik, S.W. | | Deposit date: | 1996-04-04 | | Release date: | 1997-04-21 | | Last modified: | 2022-02-23 | | Method: | SOLUTION NMR | | Cite: | X-ray and NMR structure of human Bcl-xL, an inhibitor of programmed cell death.
Nature, 381, 1996
|
|
1MAZ
 
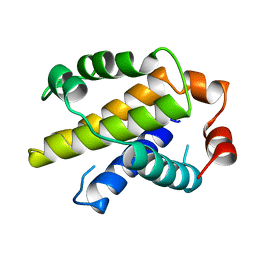 | | X-RAY STRUCTURE OF BCL-XL, AN INHIBITOR OF PROGRAMMED CELL DEATH | | Descriptor: | Bcl-2-like protein 1 | | Authors: | Muchmore, S.W, Sattler, M, Liang, H, Meadows, R.P, Harlan, J.E, Yoon, H.S, Nettesheim, D, Chang, B.S, Thompson, C.B, Wong, S.L, Ng, S.C, Fesik, S.W. | | Deposit date: | 1996-04-09 | | Release date: | 1997-04-21 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | X-ray and NMR structure of human Bcl-xL, an inhibitor of programmed cell death.
Nature, 381, 1996
|
|
3GQV
 
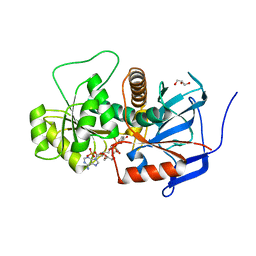 | | Lovastatin polyketide enoyl reductase (LovC) mutant K54S with bound NADP | | Descriptor: | Enoyl reductase, GLYCEROL, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Ames, B.D, Smith, P.T, Ma, S.M, Kaake, R, Wong, E.W, Wong, S.K, Xie, X, Li, J.W, Vederas, J.C, Tang, Y, Tsai, S.-C. | | Deposit date: | 2009-03-24 | | Release date: | 2009-04-07 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.74 Å) | | Cite: | biosynthesis of Lovastatin: Crystal structure and biochemical
studies of LOVC, A trans-acting polyketide enoyl reductase
To be Published
|
|
1OSS
 
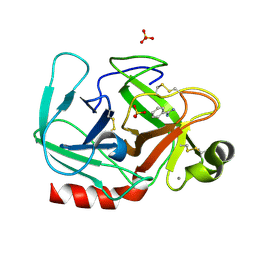 | | T190P STREPTOMYCES GRISEUS TRYPSIN IN COMPLEX WITH BENZAMIDINE | | Descriptor: | BENZAMIDINE, CALCIUM ION, SULFATE ION, ... | | Authors: | Page, M.J, Wong, S.L, Hewitt, J, Strynadka, N.C, MacGillivray, R.T. | | Deposit date: | 2003-03-20 | | Release date: | 2003-08-19 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.93 Å) | | Cite: | Engineering the Primary Substrate Specificity of Streptomyces griseus Trypsin.
Biochemistry, 42, 2003
|
|
1OS8
 
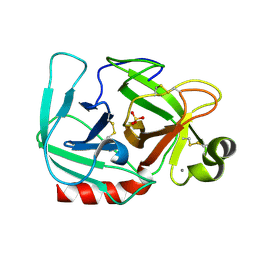 | | RECOMBINANT STREPTOMYCES GRISEUS TRYPSIN | | Descriptor: | CALCIUM ION, SULFATE ION, trypsin | | Authors: | Page, M.J, Wong, S.L, Hewitt, J, Strynadka, N.C, MacGillivray, R.T. | | Deposit date: | 2003-03-18 | | Release date: | 2003-08-19 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Engineering the Primary Substrate Specificity of Streptomyces griseus Trypsin.
Biochemistry, 42, 2003
|
|
1L8P
 
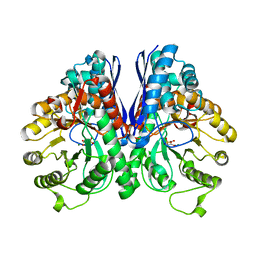 | | Mg-phosphonoacetohydroxamate complex of S39A yeast enolase 1 | | Descriptor: | MAGNESIUM ION, PHOSPHONOACETOHYDROXAMIC ACID, enolase 1 | | Authors: | Poyner, R.R, Larsen, T.M, Wong, S.W, Reed, G.H. | | Deposit date: | 2002-03-21 | | Release date: | 2002-04-03 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Functional and structural changes due to a serine to alanine mutation in the active-site flap of enolase.
Arch.Biochem.Biophys., 401, 2002
|
|
4LTP
 
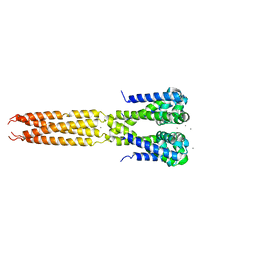 | | Bacterial sodium channel in high calcium, I222 space group, crystal 2 | | Descriptor: | CALCIUM ION, Ion transport protein | | Authors: | Shaya, D, Findeisen, F, Abderemane-Ali, F, Arrigoni, C, Wong, S, Reddy Nurva, S, Loussouarn, G, Minor, D.L. | | Deposit date: | 2013-07-23 | | Release date: | 2013-10-23 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (3.8 Å) | | Cite: | Structure of a prokaryotic sodium channel pore reveals essential gating elements and an outer ion binding site common to eukaryotic channels.
J.Mol.Biol., 426, 2014
|
|
4LTO
 
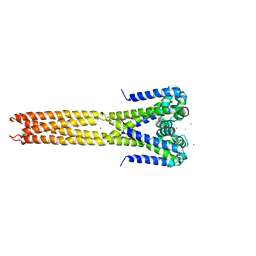 | | Bacterial sodium channel in high calcium, I222 space group | | Descriptor: | CALCIUM ION, Ion transport protein, NICKEL (II) ION | | Authors: | Shaya, D, Findeisen, F, Abderemane-Ali, F, Arrigoni, C, Wong, S, Reddy Nurva, S, Loussouarn, G, Minor, D.L. | | Deposit date: | 2013-07-23 | | Release date: | 2013-10-23 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (3.46 Å) | | Cite: | Structure of a prokaryotic sodium channel pore reveals essential gating elements and an outer ion binding site common to eukaryotic channels.
J.Mol.Biol., 426, 2014
|
|
4LTR
 
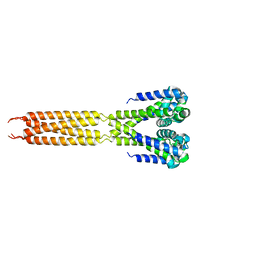 | | Bacterial sodium channel, His245Gly mutant, I222 space group | | Descriptor: | Ion transport protein | | Authors: | Shaya, D, Findeisen, F, Abderemane-Ali, F, Arrigoni, C, Wong, S, Reddy Nurva, S, Loussouarn, G, Minor, D.L. | | Deposit date: | 2013-07-23 | | Release date: | 2013-10-23 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (5.8 Å) | | Cite: | Structure of a prokaryotic sodium channel pore reveals essential gating elements and an outer ion binding site common to eukaryotic channels.
J.Mol.Biol., 426, 2014
|
|
4LTQ
 
 | | Bacterial sodium channel in low calcium, P42 space group | | Descriptor: | Ion transport protein | | Authors: | Shaya, D, Findeisen, F, Abderemane-Ali, F, Arrigoni, C, Wong, S, Reddy Nurva, S, Loussouarn, G, Minor, D.L. | | Deposit date: | 2013-07-23 | | Release date: | 2013-10-23 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (5.5 Å) | | Cite: | Structure of a prokaryotic sodium channel pore reveals essential gating elements and an outer ion binding site common to eukaryotic channels.
J.Mol.Biol., 426, 2014
|
|
2LAS
 
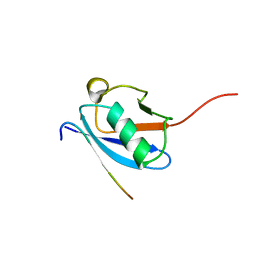 | | Molecular Determinants of Paralogue-Specific SUMO-SIM Recognition | | Descriptor: | M-IR2_peptide, Small ubiquitin-related modifier 1 | | Authors: | Namanja, A, Li, Y, Su, Y, Wong, S, Lu, J, Colson, L, Wu, C, Li, S, Chen, Y. | | Deposit date: | 2011-03-20 | | Release date: | 2011-12-14 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | Insights into High Affinity Small Ubiquitin-like Modifier (SUMO) Recognition by SUMO-interacting Motifs (SIMs) Revealed by a Combination of NMR and Peptide Array Analysis.
J.Biol.Chem., 287, 2012
|
|
