4AC3
 
 | | S.pneumoniae GlmU in complex with an antibacterial inhibitor | | Descriptor: | BIFUNCTIONAL PROTEIN GLMU, N-{2,4-DIMETHOXY-5-[(2-PIPERIDIN-1-YLBENZYL)sulfamoyl]phenyl}acetamide, SULFATE ION | | Authors: | Otterbein, L, Breed, J, Ogg, D.J. | | Deposit date: | 2011-12-12 | | Release date: | 2012-02-15 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Inhibitors of Acetyltransferase Domain of N-Acetylglucosamine-1-Phosphate-Uridyltransferase/ Glucosamine-1-Phosphate-Acetyltransferase (Glmu). Part 1: Hit to Lead Evaluation of a Novel Arylsulfonamide Series.
Bioorg.Med.Chem.Lett., 22, 2012
|
|
4AA7
 
 | | E.coli GlmU in complex with an antibacterial inhibitor | | Descriptor: | BIFUNCTIONAL PROTEIN GLMU, N-(2,4-dimethoxy-5-{[(2R)-2-methyl-2,3-dihydro-1H-indol-1-yl]sulfonyl}phenyl)acetamide, SULFATE ION | | Authors: | Otterbein, L, Breed, J, Ogg, D.J. | | Deposit date: | 2011-11-30 | | Release date: | 2012-08-15 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Inhibitors of Acetyltransferase Domain of N-Acetylglucosamine-1-Phosphate-Uridyltransferase/ Glucosamine-1-Phosphate-Acetyltransferase (Glmu). Part 1: Hit to Lead Evaluation of a Novel Arylsulfonamide Series.
Bioorg.Med.Chem.Lett., 22, 2012
|
|
4AAW
 
 | | S.pneumoniae GlmU in complex with an antibacterial inhibitor | | Descriptor: | 4-{[1-(2-{[({5-[(3-carboxypropanoyl)amino]-2,4-dimethoxyphenyl}sulfonyl)amino]methyl}phenyl)piperidin-4-yl]methoxy}-4-oxobutanoic acid, BIFUNCTIONAL PROTEIN GLMU, SULFATE ION | | Authors: | Otterbein, L, Breed, J, Ogg, D.J. | | Deposit date: | 2011-12-05 | | Release date: | 2012-02-15 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Inhibitors of Acetyltransferase Domain of N-Acetylglucosamine-1-Phosphate-Uridyltransferase/ Glucosamine-1-Phosphate-Acetyltransferase (Glmu). Part 1: Hit to Lead Evaluation of a Novel Arylsulfonamide Series.
Bioorg.Med.Chem.Lett., 22, 2012
|
|
5KO1
 
 | | Pseudokinase Domain of MLKL bound to Compound 4. | | Descriptor: | Mixed lineage kinase domain-like protein, [(1~{R})-2-[(4-fluorophenyl)amino]-2-oxidanylidene-1-phenyl-ethyl] 3-azanylpyrazine-2-carboxylate | | Authors: | Marcotte, D.J. | | Deposit date: | 2016-06-29 | | Release date: | 2016-11-16 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.16 Å) | | Cite: | ATP-Competitive MLKL Binders Have No Functional Impact on Necroptosis.
Plos One, 11, 2016
|
|
5TA8
 
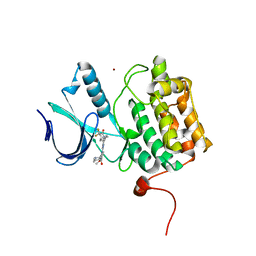 | | Crystal structure of PLK1 in complex with a novel 5,6-dihydroimidazolo[1,5-f]pteridine inhibitor | | Descriptor: | 4-[(9-cyclopentyl-7,7-difluoro-5-methyl-6-oxo-6,7,8,9-tetrahydro-5H-pyrimido[4,5-b][1,4]diazepin-2-yl)amino]-3-methoxy-N-(1-methylpiperidin-4-yl)benzamide, Serine/threonine-protein kinase PLK1, ZINC ION | | Authors: | Skene, R.J, Hosfield, D.J. | | Deposit date: | 2016-09-09 | | Release date: | 2017-02-22 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structure-based design and SAR development of 5,6-dihydroimidazolo[1,5-f]pteridine derivatives as novel Polo-like kinase-1 inhibitors.
Bioorg. Med. Chem. Lett., 27, 2017
|
|
5T1Z
 
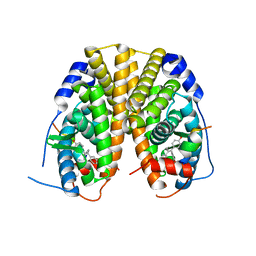 | | Estrogen Receptor Alpha Ligand Binding Domain Y537S Mutant in Complex with Ethoxytriphenylethylene and GRIP Peptide | | Descriptor: | 4,4'-[(1Z)-1-(4-ethoxyphenyl)but-1-ene-1,2-diyl]diphenol, Estrogen receptor, Nuclear receptor coactivator 2 | | Authors: | Fanning, S.W, Rajan, S.S, Maximov, P.Y, Abderrahman, B.H, Surojeet, S, Fernandes, D.J, Fan, P, Curpan, R.F, Greene, G.L, Jordan, V.C. | | Deposit date: | 2016-08-22 | | Release date: | 2017-08-30 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.102 Å) | | Cite: | Endoxifen, 4-Hydroxytamoxifen and an Estrogenic Derivative Modulate Estrogen Receptor Complex Mediated Apoptosis in Breast Cancer.
Mol. Pharmacol., 94, 2018
|
|
5TA6
 
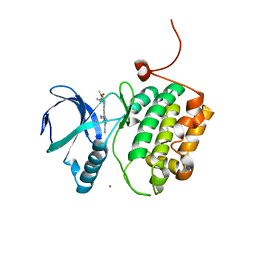 | | Crystal structure of PLK1 in complex with a novel 5,6-dihydroimidazolo[1,5-f]pteridine inhibitor. | | Descriptor: | 4-{[(6R)-7-cyano-5-cyclopentyl-6-ethyl-5,6-dihydroimidazo[1,5-f]pteridin-3-yl]amino}-3-methoxy-N-(1-methylpiperidin-4-yl)benzamide, Serine/threonine-protein kinase PLK1, ZINC ION | | Authors: | Skene, R.J, Hosfield, D.J. | | Deposit date: | 2016-09-09 | | Release date: | 2017-02-22 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structure-based design and SAR development of 5,6-dihydroimidazolo[1,5-f]pteridine derivatives as novel Polo-like kinase-1 inhibitors.
Bioorg. Med. Chem. Lett., 27, 2017
|
|
5V4W
 
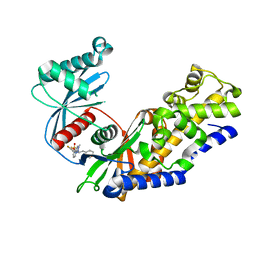 | | Human glucokinase in complex with novel indazole activator. | | Descriptor: | (2S)-2-[4-(cyclopropylsulfonyl)-1H-indazol-1-yl]-N-(5-fluoro-1,3-thiazol-2-yl)-3-(oxan-4-yl)propanamide, Glucokinase, IODIDE ION, ... | | Authors: | Skene, R.J, Hosfield, D.J. | | Deposit date: | 2017-03-10 | | Release date: | 2017-05-31 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.39 Å) | | Cite: | Discovery of potent and orally active 1,4-disubstituted indazoles as novel allosteric glucokinase activators.
Bioorg. Med. Chem. Lett., 27, 2017
|
|
5V4X
 
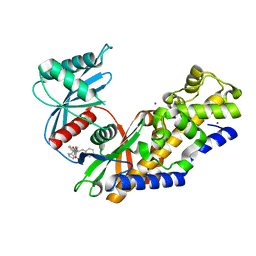 | | Human glucokinase in complex with novel pyrazole activator. | | Descriptor: | (2S)-3-cyclohexyl-2-[4-(cyclopentylsulfonyl)-2-oxopyridin-1(2H)-yl]-N-(1,3-thiazol-2-yl)propanamide, Glucokinase, IODIDE ION, ... | | Authors: | Skene, R.J, Hosfiled, D.J. | | Deposit date: | 2017-03-10 | | Release date: | 2017-05-31 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.08 Å) | | Cite: | Discovery of potent and orally active 1,4-disubstituted indazoles as novel allosteric glucokinase activators.
Bioorg. Med. Chem. Lett., 27, 2017
|
|
1QG9
 
 | | SECOND REPEAT (IS2MIC) FROM VOLTAGE-GATED SODIUM CHANNEL | | Descriptor: | PROTEIN (SODIUM CHANNEL PROTEIN, BRAIN II ALPHA SUBUNIT) | | Authors: | Doak, D.J, Mulvey, D, Kawaguchi, K, Villalain, J, Campbell, I.D. | | Deposit date: | 1999-04-22 | | Release date: | 1999-04-30 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | Structural studies of synthetic peptides dissected from the voltage-gated sodium channel.
J.Mol.Biol., 258, 1996
|
|
1MUC
 
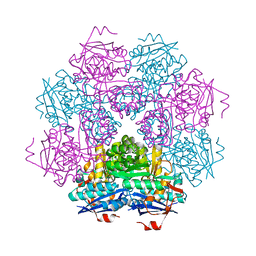 | | STRUCTURE OF MUCONATE LACTONIZING ENZYME AT 1.85 ANGSTROMS RESOLUTION | | Descriptor: | MANGANESE (II) ION, MUCONATE LACTONIZING ENZYME | | Authors: | Helin, S, Kahn, P.C, Guha, B.H.L, Mallows, D.J, Goldman, A. | | Deposit date: | 1995-09-20 | | Release date: | 1996-07-11 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | The refined X-ray structure of muconate lactonizing enzyme from Pseudomonas putida PRS2000 at 1.85 A resolution.
J.Mol.Biol., 254, 1995
|
|
1F8Z
 
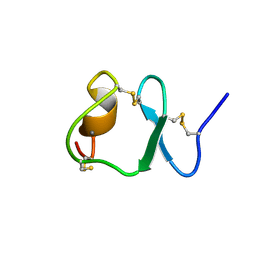 | | NMR STRUCTURE OF THE SIXTH LIGAND-BINDING MODULE OF THE LDL RECEPTOR | | Descriptor: | CALCIUM ION, LOW-DENSITY LIPOPROTEIN RECEPTOR | | Authors: | Clayton, D.J, Brereton, I.M, Kroon, P.A, Smith, R. | | Deposit date: | 2000-07-05 | | Release date: | 2000-10-18 | | Last modified: | 2022-02-23 | | Method: | SOLUTION NMR | | Cite: | Three-dimensional NMR structure of the sixth ligand-binding module of the human LDL receptor: comparison of two adjacent modules with different ligand binding specificities.
FEBS Lett., 479, 2000
|
|
4R0E
 
 | |
4QUC
 
 | | Crystal structure of chromodomain of Rhino | | Descriptor: | RE36324p | | Authors: | Li, S, Patel, D.J. | | Deposit date: | 2014-07-10 | | Release date: | 2014-08-20 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.502 Å) | | Cite: | Transgenerationally inherited piRNAs trigger piRNA biogenesis by changing the chromatin of piRNA clusters and inducing precursor processing.
Genes Dev., 28, 2014
|
|
4QUF
 
 | | crystal structure of chromodomain of Rhino with H3K9me3 | | Descriptor: | H3(1-15)K9me3 peptide, RE36324p | | Authors: | Li, S, Patel, D.J. | | Deposit date: | 2014-07-10 | | Release date: | 2014-08-20 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.501 Å) | | Cite: | Transgenerationally inherited piRNAs trigger piRNA biogenesis by changing the chromatin of piRNA clusters and inducing precursor processing.
Genes Dev., 28, 2014
|
|
4V6T
 
 | | Structure of the bacterial ribosome complexed by tmRNA-SmpB and EF-G during translocation and MLD-loading | | Descriptor: | 16S ribosomal RNA, 23S ribosomal RNA, 30S ribosomal protein S10, ... | | Authors: | Ramrath, D.J.F, Yamamoto, H, Rother, K, Wittek, D, Pech, M, Mielke, T, Loerke, J, Scheerer, P, Ivanov, P, Teraoka, Y, Shpanchenko, O, Nierhaus, K.H, Spahn, C.M.T. | | Deposit date: | 2012-01-27 | | Release date: | 2014-07-09 | | Last modified: | 2024-02-28 | | Method: | ELECTRON MICROSCOPY (8.3 Å) | | Cite: | The complex of tmRNA-SmpB and EF-G on translocating ribosomes.
Nature, 485, 2012
|
|
2C7H
 
 | | Solution NMR structure of the DWNN domain from human RBBP6 | | Descriptor: | RETINOBLASTOMA-BINDING PROTEIN 6, ISOFORM 3 | | Authors: | Pugh, D.J.R, Ab, E, Faro, A, Lutya, P.T, Hoffmann, E, Rees, D.J.G. | | Deposit date: | 2005-11-24 | | Release date: | 2006-01-19 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Dwnn, a Novel Ubiquitin-Like Domain, Implicates Rbbp6 in Mrna Processing and Ubiquitin-Like Pathways
Bmc Struct.Biol., 6, 2006
|
|
4EB1
 
 | | Hyperstable in-frame insertion variant of antithrombin | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Antithrombin-III | | Authors: | Martinez-Martinez, I, Johnson, D.J.D, Yamasaki, M, Corral, J, Huntington, J.A. | | Deposit date: | 2012-03-23 | | Release date: | 2012-07-18 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Type II antithrombin deficiency caused by a large in-frame insertion: structural, functional and pathological relevance.
J.Thromb.Haemost., 10, 2012
|
|
7ZXY
 
 | | 3.15 Angstrom cryo-EM structure of the dimeric cytochrome b6f complex from Synechocystis sp. PCC 6803 with natively bound plastoquinone and lipid molecules. | | Descriptor: | (1R)-2-{[{[(2S)-2,3-DIHYDROXYPROPYL]OXY}(HYDROXY)PHOSPHORYL]OXY}-1-[(PALMITOYLOXY)METHYL]ETHYL (11E)-OCTADEC-11-ENOATE, CHLOROPHYLL A, Cytochrome B6, ... | | Authors: | Malone, L.A, Procter, M.S, Farmer, D.F, Swainsbury, D.J.K, Hawkings, F.R, Pastorelli, F, Emrich-Mills, T.Z, Siebert, A, Hunter, C.N, Hitchcock, A, Johnson, M.P. | | Deposit date: | 2022-05-23 | | Release date: | 2022-07-06 | | Last modified: | 2022-07-27 | | Method: | ELECTRON MICROSCOPY (3.15 Å) | | Cite: | Cryo-EM structures of the Synechocystis sp. PCC 6803 cytochrome b6f complex with and without the regulatory PetP subunit.
Biochem.J., 479, 2022
|
|
8ASJ
 
 | | Four subunit cytochrome b-c1 complex from Rhodobacter sphaeroides in native nanodiscs - focussed refinement in the b-c conformation | | Descriptor: | 1,2-dioleoyl-sn-glycero-3-phosphoethanolamine, Cytochrome b, Cytochrome b-c1 subunit IV, ... | | Authors: | Swainsbury, D.J.K, Hawkings, F.R, Martin, E.C, Musial, S, Salisbury, J.H, Jackson, P.J, Farmer, D.A, Johnson, M.P, Siebert, C.A, Hitchcock, A, Hunter, C.N. | | Deposit date: | 2022-08-19 | | Release date: | 2023-03-15 | | Last modified: | 2023-03-22 | | Method: | ELECTRON MICROSCOPY (3.75 Å) | | Cite: | Cryo-EM structure of the four-subunit Rhodobacter sphaeroides cytochrome bc 1 complex in styrene maleic acid nanodiscs.
Proc.Natl.Acad.Sci.USA, 120, 2023
|
|
8ASI
 
 | | Four subunit cytochrome b-c1 complex from Rhodobacter sphaeroides in native nanodiscs - consensus refinement in the b-b conformation | | Descriptor: | 1,2-dioleoyl-sn-glycero-3-phosphoethanolamine, Cytochrome b, Cytochrome b-c1 subunit IV, ... | | Authors: | Swainsbury, D.J.K, Hawkings, F.R, Martin, E.C, Musial, S, Salisbury, J.H, Jackson, P.J, Farmer, D.A, Johnson, M.P, Siebert, C.A, Hitchcock, A, Hunter, C.N. | | Deposit date: | 2022-08-19 | | Release date: | 2023-03-15 | | Last modified: | 2023-03-22 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Cryo-EM structure of the four-subunit Rhodobacter sphaeroides cytochrome bc 1 complex in styrene maleic acid nanodiscs.
Proc.Natl.Acad.Sci.USA, 120, 2023
|
|
8HIO
 
 | | Cryo-EM structure of the Cas12m2-crRNA binary complex | | Descriptor: | Cas12m2, MAGNESIUM ION, RNA (56-MER), ... | | Authors: | Omura, N.S, Nakagawa, R, Wu, Y.W, Sudfeld, C, Warren, V.R, Hirano, H, Kusakizako, T, Kise, Y, Lebbink, H.G.J, Itoh, Y, Oost, V.D.J, Nureki, O. | | Deposit date: | 2022-11-21 | | Release date: | 2023-04-12 | | Last modified: | 2023-08-30 | | Method: | ELECTRON MICROSCOPY (3.73 Å) | | Cite: | Mechanistic and evolutionary insights into a type V-M CRISPR-Cas effector enzyme.
Nat.Struct.Mol.Biol., 30, 2023
|
|
8HHM
 
 | | Cryo-EM structure of the Cas12m2-crRNA-target DNA ternary complex intermediate state | | Descriptor: | Cas12m2, DNA (36-MER), MAGNESIUM ION, ... | | Authors: | Omura, N.S, Nakagawa, R, Wu, Y.W, Sudfeld, C, Warren, V.R, Hirano, H, Kusakizako, T, Kise, Y, Lebbink, H.G.J, Itoh, Y, Oost, V.D.J, Nureki, O. | | Deposit date: | 2022-11-16 | | Release date: | 2023-04-12 | | Last modified: | 2024-05-22 | | Method: | ELECTRON MICROSCOPY (3.08 Å) | | Cite: | Mechanistic and evolutionary insights into a type V-M CRISPR-Cas effector enzyme.
Nat.Struct.Mol.Biol., 30, 2023
|
|
8HHL
 
 | | Cryo-EM structure of the Cas12m2-crRNA-target DNA full R-loop complex | | Descriptor: | Cas12m2, MAGNESIUM ION, NTS (36-MER), ... | | Authors: | Omura, N.S, Nakagawa, R, Wu, Y.W, Sudfeld, C, Warren, V.R, Hirano, H, Kusakizako, T, Kise, Y, Lebbink, H.G.J, Itoh, Y, Oost, V.D.J, Nureki, O. | | Deposit date: | 2022-11-16 | | Release date: | 2023-04-12 | | Last modified: | 2023-08-30 | | Method: | ELECTRON MICROSCOPY (2.87 Å) | | Cite: | Mechanistic and evolutionary insights into a type V-M CRISPR-Cas effector enzyme.
Nat.Struct.Mol.Biol., 30, 2023
|
|
7EL5
 
 | |
