1CVX
 
 | | CRYSTAL STRUCTURE OF POLYAMIDE DIMER (IMPYHPPYBETADP)2 BOUND TO B-DNA DECAMER CCAGATCTGG | | Descriptor: | 5'-D(*CP*CP*AP*GP*AP*TP*CP*TP*GP*G)-3', HYDROXYPYRROLE-IMIDAZOLE-PYRROLE POLYAMIDE | | Authors: | Kielkopf, C.L, Bremer, R.E, White, S, Baird, E.E, Dervan, P.B, Rees, D.C. | | Deposit date: | 1999-08-24 | | Release date: | 2000-01-15 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.27 Å) | | Cite: | Structural effects of DNA sequence on T.A recognition by hydroxypyrrole/pyrrole pairs in the minor groove.
J.Mol.Biol., 295, 2000
|
|
1CXA
 
 | | CRYSTALLIZATION AND X-RAY STRUCTURE DETERMINATION OF CYTOCHROME C2 FROM RHODOBACTER SPHAEROIDES IN THREE CRYSTAL FORMS | | Descriptor: | CYTOCHROME C2, HEME C, IMIDAZOLE | | Authors: | Axelrod, H.L, Feher, G, Allen, J.P, Chirino, A.J, Day, M.W, Hsu, B.T, Rees, D.C. | | Deposit date: | 1994-02-14 | | Release date: | 1995-07-10 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystallization and X-ray structure determination of cytochrome c2 from Rhodobacter sphaeroides in three crystal forms.
Acta Crystallogr.,Sect.D, 50, 1994
|
|
1CVU
 
 | | CRYSTAL STRUCTURE OF ARACHIDONIC ACID BOUND TO THE CYCLOOXYGENASE ACTIVE SITE OF COX-2 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ARACHIDONIC ACID, ... | | Authors: | Kiefer, J.R, Pawlitz, J.L, Moreland, K.T, Stegeman, R.A, Gierse, J.K, Stevens, A.M, Goodwin, D.C, Rowlinson, S.W, Marnett, L.J, Stallings, W.C, Kurumbail, R.G. | | Deposit date: | 1999-08-24 | | Release date: | 2000-05-16 | | Last modified: | 2021-11-03 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural insights into the stereochemistry of the cyclooxygenase reaction.
Nature, 405, 2000
|
|
1CQ3
 
 | | STRUCTURE OF A SOLUBLE SECRETED CHEMOKINE INHIBITOR, VCCI, FROM COWPOX VIRUS | | Descriptor: | VIRAL CHEMOKINE INHIBITOR | | Authors: | Carfi, A, Smith, C.A, Smolak, P.J, McGrew, J, Wiley, D.C. | | Deposit date: | 1999-08-05 | | Release date: | 1999-11-12 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Structure of a soluble secreted chemokine inhibitor vCCI (p35) from cowpox virus.
Proc.Natl.Acad.Sci.USA, 96, 1999
|
|
7RFO
 
 | | SeMet Tailspike protein 4 (TSP4) phage CBA120, residues 1-335, obtained in the presence of LiSO4 | | Descriptor: | Tailspike protein | | Authors: | Chao, K, Shang, X, Grenfield, J, Linden, S.B, Nelson, D.C, Herzberg, O. | | Deposit date: | 2021-07-14 | | Release date: | 2022-02-23 | | Method: | X-RAY DIFFRACTION (3.02 Å) | | Cite: | Structure of Escherichia coli O157:H7 bacteriophage CBA120 tailspike protein 4 baseplate anchor and tailspike assembly domains (TSP4-N).
Sci Rep, 12, 2022
|
|
7REJ
 
 | | Tailspike protein 4 (TSP4) from phage CBA120, residues 1-335, obtained in the presence of NaK-Tartrate | | Descriptor: | IMIDAZOLE, Tailspike protein | | Authors: | Chao, K, Shang, X, Grenfield, J, Linden, S.B, Nelson, D.C, Herzberg, O. | | Deposit date: | 2021-07-13 | | Release date: | 2022-02-23 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structure of Escherichia coli O157:H7 bacteriophage CBA120 tailspike protein 4 baseplate anchor and tailspike assembly domains (TSP4-N).
Sci Rep, 12, 2022
|
|
7RFV
 
 | | Tailspike protein 4 (TSP4) from phage CBA120, residues 1-250, obtained in the presence of PEG8000 | | Descriptor: | Tailspike protein | | Authors: | Chao, K, Shang, X, Grenfield, J, Linden, S.B, Nelson, D.C, Herzberg, O. | | Deposit date: | 2021-07-14 | | Release date: | 2022-02-23 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Structure of Escherichia coli O157:H7 bacteriophage CBA120 tailspike protein 4 baseplate anchor and tailspike assembly domains (TSP4-N).
Sci Rep, 12, 2022
|
|
1IIE
 
 | |
6RXI
 
 | | In-flow serial synchrotron crystallography using a 3D-printed microfluidic device (3D-MiXD): Lysozyme | | Descriptor: | CHLORIDE ION, DI(HYDROXYETHYL)ETHER, Lysozyme C, ... | | Authors: | Monteiro, D.C.F, von Stetten, D, Pearson, A.R, Trebbin, M. | | Deposit date: | 2019-06-08 | | Release date: | 2020-03-18 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | 3D-MiXD: 3D-printed X-ray-compatible microfluidic devices for rapid, low-consumption serial synchrotron crystallography data collection in flow.
Iucrj, 7, 2020
|
|
1IM3
 
 | | Crystal Structure of the human cytomegalovirus protein US2 bound to the MHC class I molecule HLA-A2/tax | | Descriptor: | HLA CLASS I HISTOCOMPATIBILITY ANTIGEN, A-2 ALPHA CHAIN, Human T-cell lymphotropic virus type 1 Tax peptide, ... | | Authors: | Gewurz, B.E, Gaudet, R, Tortorella, D, Wang, E.W, Ploegh, H.L, Wiley, D.C. | | Deposit date: | 2001-05-09 | | Release date: | 2001-06-06 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Antigen presentation subverted: Structure of the human cytomegalovirus protein US2 bound to the class I molecule HLA-A2.
Proc.Natl.Acad.Sci.USA, 98, 2001
|
|
6SA9
 
 | |
6SXB
 
 | | XPF-ERCC1 Cryo-EM Structure, DNA-Bound form | | Descriptor: | DNA (5'-D(*TP*CP*AP*GP*CP*AP*TP*CP*TP*G)-3'), DNA (5'-D(P*CP*AP*GP*AP*TP*GP*CP*TP*GP*A)-3'), DNA excision repair protein ERCC-1, ... | | Authors: | Jones, M.L, Briggs, D.C, McDonald, N.Q. | | Deposit date: | 2019-09-25 | | Release date: | 2020-03-11 | | Last modified: | 2024-05-22 | | Method: | ELECTRON MICROSCOPY (7.9 Å) | | Cite: | Cryo-EM structures of the XPF-ERCC1 endonuclease reveal how DNA-junction engagement disrupts an auto-inhibited conformation.
Nat Commun, 11, 2020
|
|
6SXA
 
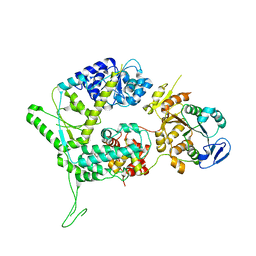 | | XPF-ERCC1 Cryo-EM Structure, Apo-form | | Descriptor: | DNA excision repair protein ERCC-1, DNA repair endonuclease XPF | | Authors: | Jones, M.L, Briggs, D.C, McDonald, N.Q. | | Deposit date: | 2019-09-25 | | Release date: | 2020-03-11 | | Last modified: | 2024-05-22 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Cryo-EM structures of the XPF-ERCC1 endonuclease reveal how DNA-junction engagement disrupts an auto-inhibited conformation.
Nat Commun, 11, 2020
|
|
6RXH
 
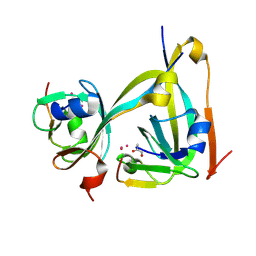 | | In-flow serial synchrotron crystallography using a 3D-printed microfluidic device (3D-MiXD): Aspartate alpha-decarboxylase | | Descriptor: | Aspartate 1-decarboxylase, UNKNOWN ATOM OR ION | | Authors: | Monteiro, D.C.F, von Stetten, D, Pearson, A.R, Trebbin, M. | | Deposit date: | 2019-06-08 | | Release date: | 2020-03-18 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | 3D-MiXD: 3D-printed X-ray-compatible microfluidic devices for rapid, low-consumption serial synchrotron crystallography data collection in flow.
Iucrj, 7, 2020
|
|
8DFC
 
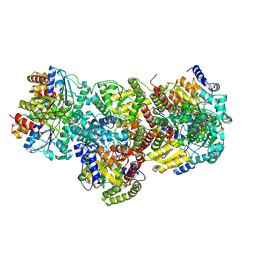 | |
8DFD
 
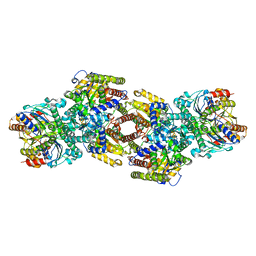 | |
7T4H
 
 | | Selenium-incorporated nitrogenase Fe protein (Av2-Se) from A. vinelandii (22 mM KSeCN, with Av1) | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Fe4-Se4 cluster, IRON/SULFUR CLUSTER, ... | | Authors: | Buscagan, T.M, Kaiser, J.T, Rees, D.C. | | Deposit date: | 2021-12-09 | | Release date: | 2022-09-14 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.51 Å) | | Cite: | Selenocyanate derived Se-incorporation into the Nitrogenase Fe protein cluster.
Elife, 11, 2022
|
|
7T3H
 
 | | MicroED structure of Dynobactin | | Descriptor: | TRP-ASN-SER-ASN-VAL-HIS-SER-TYR-ARG-PHE | | Authors: | Yoo, B.-K, Kaiser, J.T, Rees, D.C, Miller, R.D, Iinishi, A, Lewis, K, Bowman, S. | | Deposit date: | 2021-12-07 | | Release date: | 2022-10-19 | | Method: | ELECTRON CRYSTALLOGRAPHY (1.05 Å) | | Cite: | Computational identification of a systemic antibiotic for gram-negative bacteria.
Nat Microbiol, 7, 2022
|
|
6STJ
 
 | | Selective Affimers Recognize BCL-2 Family Proteins Through Non-Canonical Structural Motifs | | Descriptor: | Cystatin domain-containing protein, Induced myeloid leukemia cell differentiation protein Mcl-1 | | Authors: | Hobor, F, Miles, J.A, Trinh, C.H, Taylor, J, Tiede, C, Rowell, P.R, Jackson, B, Nadat, F, Kyle, H.F, Wicky, B.I.M, Clarke, J, Tomlinson, D.C, Wilson, A.J, Edwards, T.A. | | Deposit date: | 2019-09-10 | | Release date: | 2020-09-30 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Selective Affimers Recognise the BCL-2 Family Proteins BCL-x L and MCL-1 through Noncanonical Structural Motifs*.
Chembiochem, 22, 2021
|
|
8CH7
 
 | | RDC-refined Interleukin-4 (wild type) pH 5.6 | | Descriptor: | Interleukin-4 | | Authors: | Vaz, D.C, Rodrigues, J.R, Loureiro-Ferreira, N, Mueller, T, Sebald, W, Redfield, C, Brito, R.M.M. | | Deposit date: | 2023-02-07 | | Release date: | 2023-10-18 | | Last modified: | 2024-01-17 | | Method: | SOLUTION NMR | | Cite: | Lessons on protein structure from interleukin-4: All disulfides are not created equal.
Proteins, 92, 2024
|
|
8CGF
 
 | | Interleukin-4 (wild type) pH 2.4 | | Descriptor: | Interleukin-4 | | Authors: | Vaz, D.C, Rodrigues, J.R, Loureiro-Ferreira, N, Mueller, T, Sebald, W, Redfield, C, Brito, R.M.M. | | Deposit date: | 2023-02-04 | | Release date: | 2023-10-18 | | Last modified: | 2024-10-09 | | Method: | SOLUTION NMR | | Cite: | Lessons on protein structure from interleukin-4: All disulfides are not created equal.
Proteins, 92, 2024
|
|
8C12
 
 | | Identification of an intermediate activation state of PAK5 reveals a novel mechanism of kinase inhibition. | | Descriptor: | PAK5-Af17, Serine/threonine-protein kinase PAK 5 | | Authors: | Martin, H.L, Turner, A.L, Trinh, C.H, Bayliss, R.W, Tomlinson, D.C. | | Deposit date: | 2022-12-19 | | Release date: | 2023-10-25 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.549 Å) | | Cite: | Affimer-mediated locking of p21-activated kinase 5 in an intermediate activation state results in kinase inhibition.
Cell Rep, 42, 2023
|
|
1KFY
 
 | | QUINOL-FUMARATE REDUCTASE WITH QUINOL INHIBITOR 2-[1-(4-CHLORO-PHENYL)-ETHYL]-4,6-DINITRO-PHENOL | | Descriptor: | 2-[1-(4-CHLORO-PHENYL)-ETHYL]-4,6-DINITRO-PHENOL, FE2/S2 (INORGANIC) CLUSTER, FE3-S4 CLUSTER, ... | | Authors: | Iverson, T.M, Luna-Chavez, C, Croal, L.R, Cecchini, G, Rees, D.C. | | Deposit date: | 2001-11-24 | | Release date: | 2002-03-13 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (3.6 Å) | | Cite: | Crystallographic studies of the Escherichia coli quinol-fumarate reductase with inhibitors bound to the
quinol-binding site.
J.Biol.Chem., 277, 2002
|
|
6ST2
 
 | | Selective Affimers Recognize BCL-2 Family Proteins Through Non-Canonical Structural Motifs | | Descriptor: | Affimer AF6, Bcl-2-like protein 1, SULFATE ION | | Authors: | Hobor, F, Miles, J.A, Trinh, C.H, Taylor, J, Tiede, C, Rowell, P.R, Jackson, B, Nadat, F, Kyle, H.F, Wicky, B.I.M, Clarke, J, Tomlinson, D.C, Wilson, A.J, Edwards, T.A. | | Deposit date: | 2019-09-09 | | Release date: | 2020-09-30 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.79 Å) | | Cite: | Selective Affimers Recognise the BCL-2 Family Proteins BCL-x L and MCL-1 through Noncanonical Structural Motifs*.
Chembiochem, 22, 2021
|
|
8CRS
 
 | |
