5GPE
 
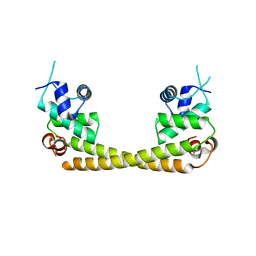 | | Crystal structure of the transcription regulator PbrR691 from Ralstonia metallidurans CH34 in complex with Lead(II) | | Descriptor: | LEAD (II) ION, Transcriptional regulator, MerR-family | | Authors: | Huang, S.Q, Chen, W.Z, Wang, D, Hu, Q.Y, Liu, X.C, Gan, J.H, Chen, H. | | Deposit date: | 2016-08-01 | | Release date: | 2016-12-28 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.01 Å) | | Cite: | Structural Basis for the Selective Pb(II) Recognition of Metalloregulatory Protein PbrR691
Inorg Chem, 55, 2016
|
|
3NL6
 
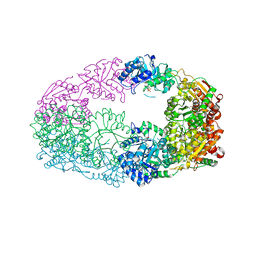 | | The Crystal Structure of Candida glabrata THI6, a Bifunctional Enzyme involved in Thiamin Biosyhthesis of Eukaryotes | | Descriptor: | MAGNESIUM ION, PHOSPHOMETHYLPHOSPHONIC ACID ADENYLATE ESTER, THIAMIN PHOSPHATE, ... | | Authors: | Paul, D, Chatterjee, A, Begley, T.P, Ealick, S.E. | | Deposit date: | 2010-06-21 | | Release date: | 2010-11-10 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.612 Å) | | Cite: | Domain Organization in Candida glabrata THI6, a Bifunctional Enzyme Required for Thiamin Biosynthesis in Eukaryotes .
Biochemistry, 49, 2010
|
|
1VDG
 
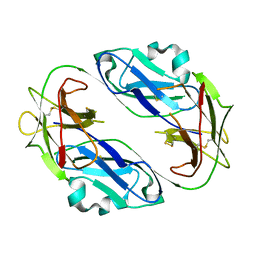 | | Crystal structure of LIR1.01, one of the alleles of LIR1 | | Descriptor: | Leukocyte immunoglobulin-like receptor subfamily B member 1 | | Authors: | Shiroishi, M, Rasubala, L, Kuroki, K, Amano, K, Tsuchiya, N, Tokunaga, K, Kohda, D, Maenaka, K. | | Deposit date: | 2004-03-22 | | Release date: | 2005-08-02 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal structure of LIR1.03, one of the alleles of LIR1
To be Published
|
|
5GSZ
 
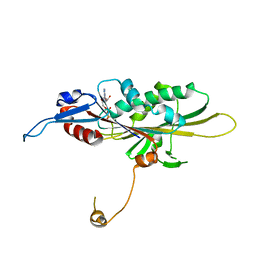 | | Crystal Structure of the KIF19A Motor Domain Complexed with Mg-ADP | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Kinesin-like protein KIF19, MAGNESIUM ION | | Authors: | Wang, D, Nitta, R, Hirokawa, N. | | Deposit date: | 2016-08-18 | | Release date: | 2016-09-28 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.72 Å) | | Cite: | Motility and microtubule depolymerization mechanisms of the Kinesin-8 motor, KIF19A
Elife, 5, 2016
|
|
3LBD
 
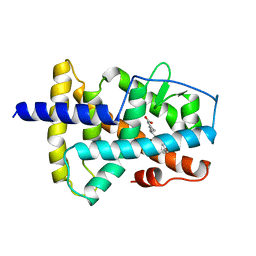 | | LIGAND-BINDING DOMAIN OF THE HUMAN RETINOIC ACID RECEPTOR GAMMA BOUND TO 9-CIS RETINOIC ACID | | Descriptor: | (9cis)-retinoic acid, RETINOIC ACID RECEPTOR GAMMA | | Authors: | Klaholz, B.P, Renaud, J.-P, Mitschler, A, Moras, D, Structural Proteomics in Europe (SPINE) | | Deposit date: | 1998-02-04 | | Release date: | 1999-03-02 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Conformational adaptation of agonists to the human nuclear receptor RAR gamma.
Nat.Struct.Biol., 5, 1998
|
|
6QVT
 
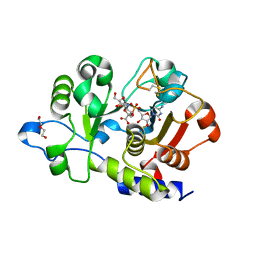 | | CMP-Sialic acid bound structure of the human wild type Beta-galactoside alpha-2,6-sialyltransferase 1 (ST6Gal1) | | Descriptor: | Beta-galactoside alpha-2,6-sialyltransferase 1, CYTIDINE-5'-MONOPHOSPHATE-5-N-ACETYLNEURAMINIC ACID, DI(HYDROXYETHYL)ETHER | | Authors: | Harrus, D, Glumoff, T. | | Deposit date: | 2019-03-04 | | Release date: | 2020-06-10 | | Last modified: | 2024-08-07 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Unliganded and CMP-Neu5Ac bound structures of human alpha-2,6-sialyltransferase ST6Gal I at high resolution.
J.Struct.Biol., 212, 2020
|
|
3EG5
 
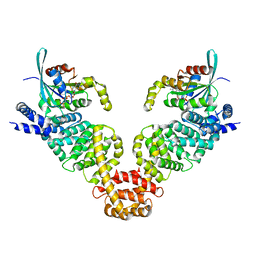 | | Crystal structure of MDIA1-TSH GBD-FH3 in complex with CDC42-GMPPNP | | Descriptor: | Cell division control protein 42 homolog, MAGNESIUM ION, PHOSPHOAMINOPHOSPHONIC ACID-GUANYLATE ESTER, ... | | Authors: | Lammers, M, Meyer, S, Kuehlmann, D, Wittinghofer, A. | | Deposit date: | 2008-09-10 | | Release date: | 2008-10-14 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Specificity of Interactions between mDia Isoforms and Rho Proteins
J.Biol.Chem., 283, 2008
|
|
1VKQ
 
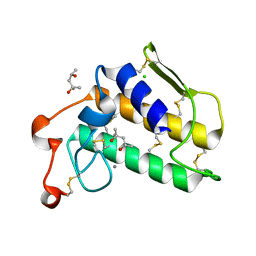 | | A re-determination of the structure of the triple mutant (K53,56,120M) of phospholipase A2 at 1.6A resolution using sulphur-SAS at 1.54A wavelength | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, CALCIUM ION, CHLORIDE ION, ... | | Authors: | Sekar, K, Velmurugan, D, Rajakannan, V, Yamane, T, Dauter, M, Dauter, Z. | | Deposit date: | 2004-06-12 | | Release date: | 2004-08-31 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | A redetermination of the structure of the triple mutant (K53,56,120M) of phospholipase A2 at 1.6 A resolution using sulfur-SAS at 1.54 A wavelength.
Acta Crystallogr.,Sect.D, 60, 2004
|
|
5GWD
 
 | | Structure of Myroilysin | | Descriptor: | Myroilysin, ZINC ION | | Authors: | Xu, D, Ran, T, Wang, W. | | Deposit date: | 2016-09-10 | | Release date: | 2017-02-15 | | Last modified: | 2017-10-18 | | Method: | X-RAY DIFFRACTION (1.89 Å) | | Cite: | Myroilysin Is a New Bacterial Member of the M12A Family of Metzincin Metallopeptidases and Is Activated by a Cysteine Switch Mechanism.
J. Biol. Chem., 292, 2017
|
|
4NHR
 
 | |
5A29
 
 | | Family 2 Pectate Lyase from Vibrio vulnificus | | Descriptor: | 1,2-ETHANEDIOL, EXOPOLYGALACTURONATE LYASE, MANGANESE (II) ION, ... | | Authors: | McLean, R, Hobbs, J.K, Suits, M.D, Tuomivaara, S, Jones, D, Boraston, A.B, Abbott, D.W. | | Deposit date: | 2015-05-15 | | Release date: | 2015-07-01 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Functional Analyses of Resurrected and Contemporary Enzymes Illuminate an Evolutionary Path for the Emergence of Exolysis in Polysaccharide Lyase Family 2.
J.Biol.Chem., 290, 2015
|
|
1UXB
 
 | | ADENOVIRUS AD19p FIBRE HEAD in complex with sialyl-lactose | | Descriptor: | ACETATE ION, FIBER PROTEIN, N-acetyl-alpha-neuraminic acid-(2-3)-beta-D-galactopyranose, ... | | Authors: | Burmeister, W.P, Guilligay, D, Cusack, S, Wadell, G, Arnberg, N. | | Deposit date: | 2004-02-24 | | Release date: | 2004-07-01 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Crystal Structure of Species D Adenovirus Fiber Knobs and Their Sialic Acid Binding Sites
J.Virol., 78, 2004
|
|
5A2J
 
 | | Crystal structure of scFv-SM3 in complex with the naked peptide APDTRP | | Descriptor: | 1,2-ETHANEDIOL, SCFV-SM3, THE NAKED PEPTIDE APDTRP | | Authors: | Martinez-Saez, N, Castro-Lopez, J, Valero-Gonzalez, J, Madariaga, D, Companon, I, Somovilla, V.J, Salvado, M, Asensio, J.L, Jimenez-Barbero, J, Avenoza, A, Busto, J.H, Bernardes, G.J.L, Peregrina, J.M, Hurtado-Guerrero, R, Corzana, F. | | Deposit date: | 2015-05-20 | | Release date: | 2015-06-03 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Deciphering the Non-Equivalence of Serine and Threonine O-Glycosylation Points: Implications for Molecular Recognition of the Tn Antigen by an Anti-Muc1 Antibody.
Angew.Chem.Int.Ed.Engl., 54, 2015
|
|
5AG5
 
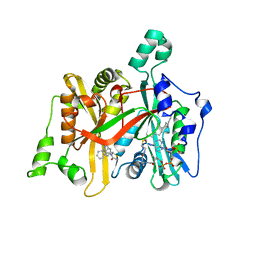 | | CRYSTAL STRUCTURE OF LEISHMANIA MAJOR N-MYRISTOYLTRANSFERASE (NMT) WITH BOUND MYRISTOYL-COA AND A THIAZOLIDINONE LIGAND | | Descriptor: | (2R)-3-benzyl-2-(1H-indazol-5-yl)-1,3-thiazolidin-4-one, GLYCYLPEPTIDE N-TETRADECANOYLTRANSFERASE, TETRADECANOYL-COA | | Authors: | Robinson, D.A, Spinks, D, Smith, V.C, Thompson, S, Smith, A, Torrie, L.S, McElroy, S.P, Brand, S, Brenk, R, Frearson, J.A, Read, K.D, Wyatt, P.G, Gilbert, I.H. | | Deposit date: | 2015-01-29 | | Release date: | 2015-10-07 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Development of Small-Molecule Trypanosoma Brucei N-Myristoyltransferase Inhibitors: Discovery and Optimisation of a Novel Binding Mode.
Chemmedchem, 10, 2015
|
|
5AKT
 
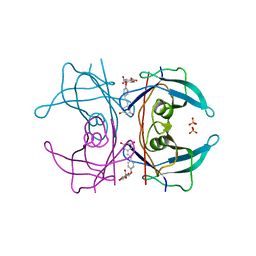 | | Transthyretin binding heterogeneity and anti-amyloidogenic activity of natural polyphenols and their metabolites: resveratrol-4'-O- glucuronide | | Descriptor: | Resveratrol-4'-O-glucuronide, SULFATE ION, TRANSTHYRETIN | | Authors: | Florio, P, Folli, C, Cianci, M, Del Rio, D, Zanotti, G, Berni, R. | | Deposit date: | 2015-03-05 | | Release date: | 2015-10-21 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Transthyretin Binding Heterogeneity and Anti-Amyloidogenic Activity of Natural Polyphenols and Their Metabolites
J.Biol.Chem., 290, 2015
|
|
5A5E
 
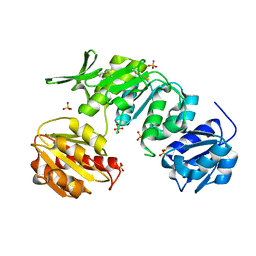 | | CRYSTAL STRUCTURE OF MURD LIGASE FROM ESCHERICHIA COLI | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, NICKEL (II) ION, SULFATE ION, ... | | Authors: | Sink, R, Kotnik, M, Zega, A, Barreteau, H, Gobec, S, Blanot, D, Dessen, A, Contreras-Martel, C. | | Deposit date: | 2015-06-17 | | Release date: | 2016-04-13 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.84 Å) | | Cite: | Crystallographic Study of Peptidoglycan Biosynthesis Enzyme MurD: Domain Movement Revisited.
PLoS ONE, 11, 2016
|
|
1USX
 
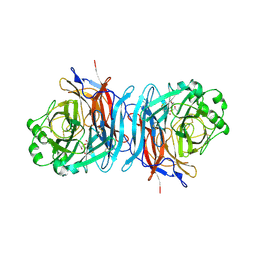 | | Crystal structure of the Newcastle disease virus hemagglutinin-neuraminidase complexed with thiosialoside | | Descriptor: | 2-DEOXY-2,3-DEHYDRO-N-ACETYL-NEURAMINIC ACID, HEMAGGLUTININ-NEURAMINIDASE GLYCOPROTEIN, N-acetyl-alpha-neuraminic acid-(2-6)-methyl 6-thio-beta-D-galactopyranoside | | Authors: | Zaitsev, V, Itzstein, M, Groves, D, Kiefel, M, Takimoto, T, Portner, A, Taylor, G. | | Deposit date: | 2003-12-01 | | Release date: | 2004-03-19 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Second Sialic Acid Binding Site in Newcastle Disease Virus Hemagglutinin-Neuraminidase: Implications for Fusion
J.Virol., 78, 2004
|
|
5AL0
 
 | | Transthyretin binding heterogeneity and anti-amyloidogenic activity of natural polyphenols and their metabolites: resveratrol-3-O-sulfate | | Descriptor: | DIMETHYL SULFOXIDE, RESVERATROL-3-O-SULFATE, SULFATE ION, ... | | Authors: | Florio, P, Foll, C, Cianci, M, Del Rio, D, Zanotti, G, Berni, R. | | Deposit date: | 2015-03-05 | | Release date: | 2015-10-21 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.388 Å) | | Cite: | Transthyretin Binding Heterogeneity and Anti-Amyloidogenic Activity of Natural Polyphenols and Their Metabolites
J.Biol.Chem., 290, 2015
|
|
5OOI
 
 | | STRUCTURE OF PROTEIN KINASE CK2 CATALYTIC SUBUNIT (ISOFORM CK2ALPHA') IN COMPLEX WITH THE INDENOINDOLE-TYPE INHIBITOR 4P | | Descriptor: | 1,2-ETHANEDIOL, 4-(3-methylbut-2-enoxy)-5-propan-2-yl-7,8-dihydro-6~{H}-indeno[1,2-b]indole-9,10-dione, ACETATE ION, ... | | Authors: | Hochscherf, J, Lindenblatt, D, Witulski, B, Birus, R, Aichele, D, Marminon, C, Bouaziz, Z, Le Borgne, M, Jose, J, Niefind, K. | | Deposit date: | 2017-08-07 | | Release date: | 2017-12-27 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.998 Å) | | Cite: | Unexpected Binding Mode of a Potent Indeno[1,2-b]indole-Type Inhibitor of Protein Kinase CK2 Revealed by Complex Structures with the Catalytic Subunit CK2 alpha and Its Paralog CK2 alpha '.
Pharmaceuticals (Basel), 10, 2017
|
|
5H1O
 
 | | CRISPR-associated protein | | Descriptor: | ACETATE ION, CRISPR-associated endoribonuclease Cas2 | | Authors: | Ka, D, Jeong, U, Bae, E. | | Deposit date: | 2016-10-11 | | Release date: | 2017-10-18 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Structural and dynamic insights into the role of conformational switching in the nuclease activity of the Xanthomonas albilineans Cas2 in CRISPR-mediated adaptive immunity
Struct Dyn, 4, 2017
|
|
5A27
 
 | | Leishmania major N-myristoyltransferase in complex with a chlorophenyl 1,2,4-oxadiazole inhibitor. | | Descriptor: | 5-chloranyl-N-[2-(3-methoxyphenyl)ethanimidoyl]-2-piperidin-4-yloxy-benzamide, GLYCYLPEPTIDE N-TETRADECANOYLTRANSFERASE, MAGNESIUM ION, ... | | Authors: | Rackham, M.D, Yu, Z, Brannigan, J.A, Heal, W.P, Paape, D, Barker, K.V, Wilkinson, A.J, Smith, D.F, Tate, E.W, Leatherbarrow, R.J. | | Deposit date: | 2015-05-15 | | Release date: | 2016-03-23 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.37 Å) | | Cite: | Discovery of High Affinity Inhibitors of Leishmania Donovani N-Myristoyltransferase.
Medchemcomm, 6, 2015
|
|
5OX9
 
 | | Structure of the Cyan Fluorescent Protein SCFP3A at pH 4.5 | | Descriptor: | CHLORIDE ION, Green fluorescent protein | | Authors: | Gotthard, G, von Stetten, D, Clavel, D, Noirclerc-Savoye, M, Royant, A. | | Deposit date: | 2017-09-06 | | Release date: | 2017-11-29 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.56 Å) | | Cite: | Chromophore Isomer Stabilization Is Critical to the Efficient Fluorescence of Cyan Fluorescent Proteins.
Biochemistry, 56, 2017
|
|
5GY7
 
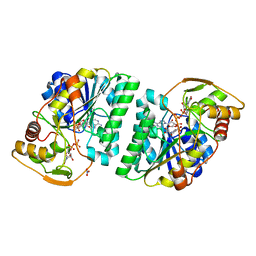 | | X-Ray structure of H243I mutant of UDP-Galactose 4-epimerase from E.coli:evidence for existence of open and closed active site during catalysis. | | Descriptor: | GLYCEROL, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, NITRATE ION, ... | | Authors: | Singh, N, Tiwari, P, Phulera, S, Dixit, A, Choudhury, D. | | Deposit date: | 2016-09-21 | | Release date: | 2016-11-30 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.43 Å) | | Cite: | X-Ray structure of H243I mutant of UDP-Galactose 4-epimerase from E.coli:evidence for existence of open and closed active site during catalysis.
To Be Published
|
|
5A9A
 
 | | Crystal structure of Simulium ubiquitum CPV20 polyhedra | | Descriptor: | POLYHEDRIN, URIDINE 5'-TRIPHOSPHATE | | Authors: | Ji, X, Axford, D, Owen, R, Evans, G, Ginn, H.M, Sutton, G, Stuart, D.I. | | Deposit date: | 2015-07-17 | | Release date: | 2015-09-02 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.822 Å) | | Cite: | Polyhedra Structures and the Evolution of the Insect Viruses.
J.Struct.Biol., 192, 2015
|
|
5H2T
 
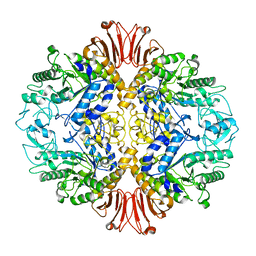 | |
