3KXC
 
 | | Mutant transport protein | | Descriptor: | PALMITIC ACID, Trafficking protein particle complex subunit 3, Trafficking protein particle complex subunit 6B | | Authors: | Kummel, D, Heinemann, U. | | Deposit date: | 2009-12-02 | | Release date: | 2010-04-21 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Characterization of the self-palmitoylation activity of the transport protein particle component Bet3
Cell.Mol.Life Sci., 67, 2010
|
|
3KPM
 
 | | Crystal Structure of HLA B*4402 in complex with EEYLKAWTF, a mimotope | | Descriptor: | Beta-2-microglobulin, EEYLKAWTF, mimotope peptide, ... | | Authors: | Macdonald, W.A, Chen, Z, Gras, S, Archbold, J.K, Tynan, F.E, Clements, C.S, Bharadwaj, M, Kjer-Nielsen, L, Saunders, P.M, Wilce, M.C, Crawford, F, Stadinsky, B, Jackson, D, Brooks, A.G, Purcell, A.W, Kappler, J.W, Burrows, S.R, Rossjohn, J, McCluskey, J. | | Deposit date: | 2009-11-16 | | Release date: | 2009-12-22 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | T cell allorecognition via molecular mimicry.
Immunity, 31, 2009
|
|
3L06
 
 | | Crystal structure of N-acetyl-L-ornithine transcarbamylase E92V mutant complexed with carbamyl phosphate and N-succinyl-L-norvaline | | Descriptor: | N-(3-CARBOXYPROPANOYL)-L-NORVALINE, N-acetylornithine carbamoyltransferase, PHOSPHORIC ACID MONO(FORMAMIDE)ESTER, ... | | Authors: | Shi, D, Yu, X, Allewell, N.M, Tuchman, M. | | Deposit date: | 2009-12-09 | | Release date: | 2010-03-31 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.81 Å) | | Cite: | A single mutation in the active site swaps the substrate specificity of N-acetyl-L-ornithine transcarbamylase and N-succinyl-L-ornithine transcarbamylase.
Protein Sci., 16, 2007
|
|
3KRZ
 
 | | Crystal Structure of the Thermostable NADH4-bound old yellow enzyme from Thermoanaerobacter pseudethanolicus E39 | | Descriptor: | 1,4,5,6-TETRAHYDRONICOTINAMIDE ADENINE DINUCLEOTIDE, FLAVIN MONONUCLEOTIDE, NADH:flavin oxidoreductase/NADH oxidase | | Authors: | Adalbjornsson, B.V, Toogood, H.S, Leys, D, Scrutton, N.S. | | Deposit date: | 2009-11-20 | | Release date: | 2009-12-08 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Biocatalysis with thermostable enzymes: structure and properties of a thermophilic 'ene'-reductase related to old yellow enzyme.
Chembiochem, 11, 2010
|
|
3KSE
 
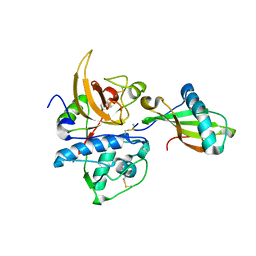 | |
3LFB
 
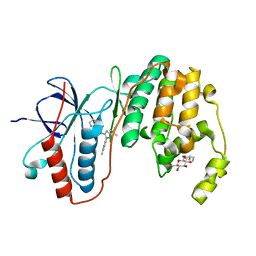 | | Human p38 MAP Kinase in Complex with RL98 | | Descriptor: | 1-[3-tert-butyl-1-(4-methylphenyl)-1H-pyrazol-5-yl]-3-(1,3-thiazol-2-yl)urea, Mitogen-activated protein kinase 14, octyl beta-D-glucopyranoside | | Authors: | Gruetter, C, Simard, J.R, Getlik, M, Rauh, D. | | Deposit date: | 2010-01-16 | | Release date: | 2011-04-20 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Development of novel thiazole-urea compounds which stabalize the inactive conformation of p38 alpha
To be Published
|
|
3LFA
 
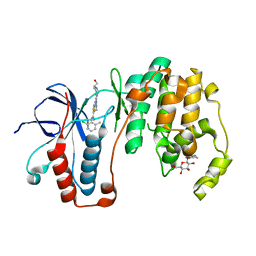 | | Human p38 MAP Kinase in Complex with Dasatinib | | Descriptor: | Mitogen-activated protein kinase 14, N-(2-CHLORO-6-METHYLPHENYL)-2-({6-[4-(2-HYDROXYETHYL)PIPERAZIN-1-YL]-2-METHYLPYRIMIDIN-4-YL}AMINO)-1,3-THIAZOLE-5-CARBOXAMIDE, octyl beta-D-glucopyranoside | | Authors: | Gruetter, C, Simard, J.R, Getlik, M, Rauh, D. | | Deposit date: | 2010-01-16 | | Release date: | 2011-04-20 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Development of novel thiazole-urea compounds which stabalize the inactive conformation of p38 alpha
To be Published
|
|
3LLY
 
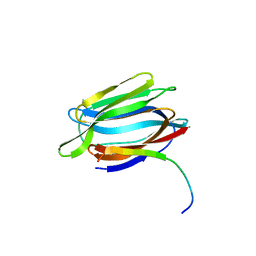 | | Crystal Structure Analysis of Maclura pomifera agglutinin | | Descriptor: | Agglutinin alpha chain, Agglutinin beta-2 chain | | Authors: | Huang, J, Xu, Z, Wang, D, Ogato, C, Hirama, T, Palczewski, K, Hazen, S.L, Lee, X, Young, N.M. | | Deposit date: | 2010-01-29 | | Release date: | 2010-09-22 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Characterization of the secondary binding sites of Maclura pomifera agglutinin by glycan array and crystallographic analyses.
Glycobiology, 20, 2010
|
|
3LLZ
 
 | | Crystal Structure Analysis of Maclura pomifera agglutinin complex with Gal-beta-1,3-GalNAc | | Descriptor: | Agglutinin alpha chain, Agglutinin beta-2 chain, beta-D-galactopyranose-(1-3)-2-acetamido-2-deoxy-beta-D-galactopyranose | | Authors: | Huang, J, Xu, Z, Wang, D, Ogato, C, Hirama, T, Palczewski, K, Hazen, S.L, Lee, X, Young, N.M. | | Deposit date: | 2010-01-29 | | Release date: | 2010-09-22 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Characterization of the secondary binding sites of Maclura pomifera agglutinin by glycan array and crystallographic analyses.
Glycobiology, 20, 2010
|
|
3LMN
 
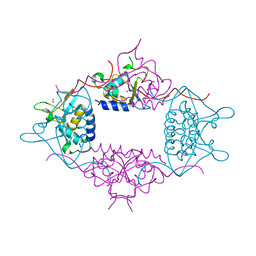 | | Oligomeric structure of the DUSP domain of human USP15 | | Descriptor: | ACETIC ACID, FORMIC ACID, Ubiquitin carboxyl-terminal hydrolase 15 | | Authors: | Walker, J.R, Asinas, A, Avvakumov, G.V, Alenkin, D, Weigelt, J, Bountra, C, Edwards, A.M, Arrowsmith, C.H, Bochkarev, A, Dhe-Paganon, S. | | Deposit date: | 2010-01-31 | | Release date: | 2010-03-23 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Crystal Structure of the Human Ubiquitin-Specific Protease 15 DUSP Domain
To be Published
|
|
3LOW
 
 | |
3LU7
 
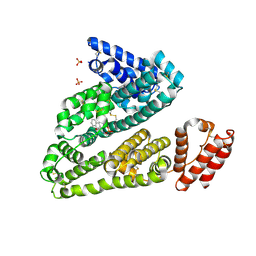 | | Human serum albumin in complex with compound 2 | | Descriptor: | 4-[(1R,2R)-2-{[(5-fluoro-1H-indol-2-yl)carbonyl]amino}-2,3-dihydro-1H-inden-1-yl]butanoic acid, PHOSPHATE ION, Serum albumin | | Authors: | Buttar, D, Colclough, N, Gerhardt, S, MacFaul, P.A, Phillips, S.D, Plowright, A, Whittamore, P, Tam, K, Maskos, K, Steinbacher, S, Steuber, H. | | Deposit date: | 2010-02-17 | | Release date: | 2010-10-27 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | A combined spectroscopic and crystallographic approach to probing drug-human serum albumin interactions
Bioorg.Med.Chem., 18, 2010
|
|
3LC1
 
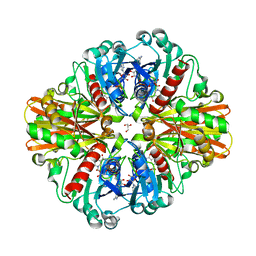 | | Crystal Structure of H178N mutant of Glyceraldehyde-3-phosphate-dehydrogenase 1 (GAPDH 1) from Staphylococcus aureus MRSA252 complexed with NAD at 2.0 angstrom resolution. | | Descriptor: | GLYCEROL, Glyceraldehyde-3-phosphate dehydrogenase 1, NICOTINAMIDE-ADENINE-DINUCLEOTIDE | | Authors: | Mukherjee, S, Dutta, D, Saha, B, Das, A.K. | | Deposit date: | 2010-01-09 | | Release date: | 2010-08-18 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of glyceraldehyde-3-phosphate dehydrogenase 1 from methicillin-resistant Staphylococcus aureus MRSA252 provides novel insights into substrate binding and catalytic mechanism.
J.Mol.Biol., 401, 2010
|
|
3LFD
 
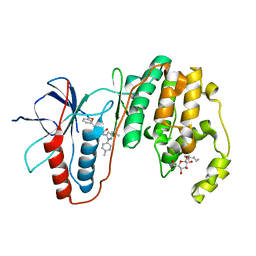 | | Human p38 MAP Kinase in Complex with RL113 | | Descriptor: | 1-{4-[2-(benzyloxy)ethyl]-1,3-thiazol-2-yl}-3-[3-tert-butyl-1-(4-methylphenyl)-1H-pyrazol-5-yl]urea, Mitogen-activated protein kinase 14, octyl beta-D-glucopyranoside | | Authors: | Gruetter, C, Simard, J.R, Getlik, M, Rauh, D. | | Deposit date: | 2010-01-16 | | Release date: | 2011-04-20 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (3.4 Å) | | Cite: | Development of novel thiazole-urea compounds which stabalize the inactive conformation of p38 alpha
To be Published
|
|
3LM6
 
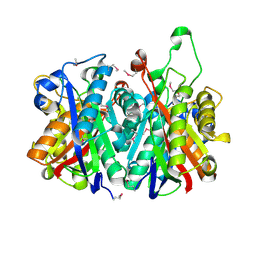 | | Crystal Structure of Stage V sporulation protein AD (spoVAD) from Bacillus subtilis, Northeast Structural Genomics Consortium Target SR525 | | Descriptor: | Stage V sporulation protein AD | | Authors: | Forouhar, F, Su, M, Seetharaman, J, Fang, F, Xiao, R, Cunningham, K, Ma, L, Wang, D, Everett, J.K, Nair, R, Acton, T.B, Rost, B, Montelione, G.T, Tong, L, Hunt, J.F, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2010-01-29 | | Release date: | 2010-02-16 | | Last modified: | 2019-07-17 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Northeast Structural Genomics Consortium Target SR525
To be Published
|
|
3LOZ
 
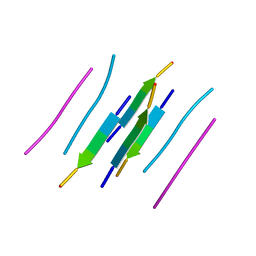 | |
3L6O
 
 | | Crystal Structure of Phosphate bound apo Glyceraldehyde-3-phosphate dehydrogenase 1 from MRSA252 at 2.2 Angstrom resolution | | Descriptor: | Glyceraldehyde-3-phosphate dehydrogenase 1, PHOSPHATE ION | | Authors: | Mukherjee, S, Dutta, D, Saha, B, Das, A.K. | | Deposit date: | 2009-12-23 | | Release date: | 2010-08-18 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structure of glyceraldehyde-3-phosphate dehydrogenase 1 from methicillin-resistant Staphylococcus aureus MRSA252 provides novel insights into substrate binding and catalytic mechanism.
J.Mol.Biol., 401, 2010
|
|
3LCC
 
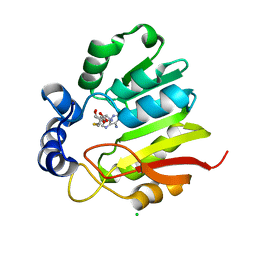 | |
5SB8
 
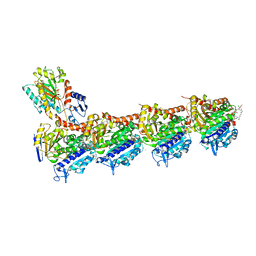 | | Tubulin-maytansinoid-3-complex | | Descriptor: | (1S,2R,3S,5S,6S,16E,18E,20R)-11-chloro-6-hydroxy-12,20-dimethoxy-2,5,9,16-tetramethyl-4,24-dioxa-9,22-diazatetracyclo[19.3.1.1~10,14~.0~3,5~]hexacosa-10(26),11,13,16,18,21-hexaene-8,23-dione, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, CALCIUM ION, ... | | Authors: | Marzullo, P, Boiarska, Z, Perez-Pena, H, Abel, A.-C, Alvarez-Bernad, B, Lucena-Agell, D, Vasile, F, Sironi, M, Steinmetz, M.O, Prota, A.E, Diaz, J.F, Pieraccini, S, Passarella, D. | | Deposit date: | 2021-07-20 | | Release date: | 2021-12-08 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Maytansinol Derivatives: Side Reactions as a Chance for New Tubulin Binders.
Chemistry, 28, 2022
|
|
5SBD
 
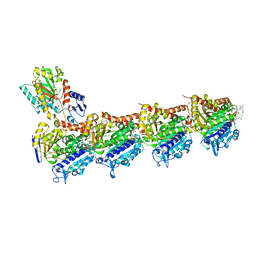 | | Tubulin-maytansinoid-5b-complex | | Descriptor: | (1S,2R,3S,5S,6S,16E,18E,20R)-11-chloro-12,20-dimethoxy-2,5,9,16-tetramethyl-8,23-dioxo-4,24-dioxa-9,22-diazatetracyclo[19.3.1.1~10,14~.0~3,5~]hexacosa-10(26),11,13,16,18,21(25)-hexaen-6-yl acetate, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, CALCIUM ION, ... | | Authors: | Marzullo, P, Boiarska, Z, Perez-Pena, H, Abel, A.-C, Alvarez-Bernad, B, Lucena-Agell, D, Vasile, F, Sironi, M, Steinmetz, M.O, Prota, A.E, Diaz, J.F, Pieraccini, S, Passarella, D. | | Deposit date: | 2021-07-20 | | Release date: | 2021-12-08 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Maytansinol Derivatives: Side Reactions as a Chance for New Tubulin Binders.
Chemistry, 28, 2022
|
|
5SBA
 
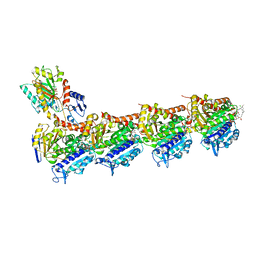 | | Tubulin-maytansinoid-4b-complex | | Descriptor: | (1S,2R,3S,5S,6S,16E,18E,20R,21S)-11-chloro-21-hydroxy-12,20-dimethoxy-2,5,9,16-tetramethyl-8,23-dioxo-4,24-dioxa-9,22-diazatetracyclo[19.3.1.1~10,14~.0~3,5~]hexacosa-10(26),11,13,16,18-pentaen-6-yl acetate, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, CALCIUM ION, ... | | Authors: | Marzullo, P, Boiarska, Z, Perez-Pena, H, Abel, A.-C, Alvarez-Bernad, B, Lucena-Agell, D, Vasile, F, Sironi, M, Steinmetz, M.O, Prota, A.E, Diaz, J.F, Pieraccini, S, Passarella, D. | | Deposit date: | 2021-07-20 | | Release date: | 2021-12-08 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Maytansinol Derivatives: Side Reactions as a Chance for New Tubulin Binders.
Chemistry, 28, 2022
|
|
5SBE
 
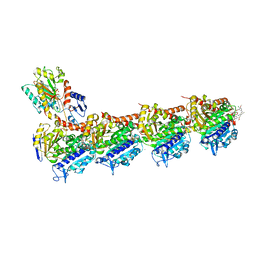 | | Tubulin-maytansinoid-5c-complex | | Descriptor: | (1S,2R,3S,5S,6S,16E,18E,20R)-11-chloro-12,20-dimethoxy-2,5,9,16-tetramethyl-8,23-dioxo-4,24-dioxa-9,22-diazatetracyclo[19.3.1.1~10,14~.0~3,5~]hexacosa-10(26),11,13,16,18,21-hexaen-6-yl hept-6-ynoate, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, CALCIUM ION, ... | | Authors: | Marzullo, P, Boiarska, Z, Perez-Pena, H, Abel, A.-C, Alvarez-Bernad, B, Lucena-Agell, D, Vasile, F, Sironi, M, Steinmetz, M.O, Prota, A.E, Diaz, J.F, Pieraccini, S, Passarella, D. | | Deposit date: | 2021-07-20 | | Release date: | 2021-12-08 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Maytansinol Derivatives: Side Reactions as a Chance for New Tubulin Binders.
Chemistry, 28, 2022
|
|
5SBB
 
 | | Tubulin-maytansinoid-4c-complex | | Descriptor: | (1R,2R,3S,5S,6S,16E,18E,20R,21S)-11-chloro-21-hydroxy-12,20-dimethoxy-2,5,9,16-tetramethyl-8,23-dioxo-4,24-dioxa-9,22-diazatetracyclo[19.3.1.1~10,14~.0~3,5~]hexacosa-10(26),11,13,16,18-pentaen-6-yl heptanoate, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, CALCIUM ION, ... | | Authors: | Marzullo, P, Boiarska, Z, Perez-Pena, H, Abel, A.-C, Alvarez-Bernad, B, Lucena-Agell, D, Vasile, F, Sironi, M, Steinmetz, M.O, Prota, A.E, Diaz, J.F, Pieraccini, S, Passarella, D. | | Deposit date: | 2021-07-20 | | Release date: | 2021-12-08 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Maytansinol Derivatives: Side Reactions as a Chance for New Tubulin Binders.
Chemistry, 28, 2022
|
|
5SBC
 
 | | Tubulin-maytansinoid-5a-complex | | Descriptor: | (1S,2R,3S,5S,6S,16E,18E,20R)-11-chloro-12,20-dimethoxy-2,5,9,16-tetramethyl-8,23-dioxo-4,24-dioxa-9,22-diazatetracyclo[19.3.1.1~10,14~.0~3,5~]hexacosa-10(26),11,13,16,18,21-hexaen-6-yl phenylacetate, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, CALCIUM ION, ... | | Authors: | Marzullo, P, Boiarska, Z, Perez-Pena, H, Abel, A.-C, Alvarez-Bernad, B, Lucena-Agell, D, Vasile, F, Sironi, M, Steinmetz, M.O, Prota, A.E, Diaz, J.F, Pieraccini, S, Passarella, D. | | Deposit date: | 2021-07-20 | | Release date: | 2021-12-08 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.32 Å) | | Cite: | Maytansinol Derivatives: Side Reactions as a Chance for New Tubulin Binders.
Chemistry, 28, 2022
|
|
5SB9
 
 | | Tubulin-maytansinoid-4a-complex | | Descriptor: | (1S,2R,3S,5S,6S,16E,18E,20R,21S)-11-chloro-21-hydroxy-12,20-dimethoxy-2,5,9,16-tetramethyl-8,23-dioxo-4,24-dioxa-9,22-diazatetracyclo[19.3.1.1~10,14~.0~3,5~]hexacosa-10(26),11,13,16,18-pentaen-6-yl phenylacetate, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, CALCIUM ION, ... | | Authors: | Marzullo, P, Boiarska, Z, Perez-Pena, H, Abel, A.-C, Alvarez-Bernad, B, Lucena-Agell, D, Vasile, F, Sironi, M, Steinmetz, M.O, Prota, A.E, Diaz, J.F, Pieraccini, S, Passarella, D. | | Deposit date: | 2021-07-20 | | Release date: | 2021-12-08 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Maytansinol Derivatives: Side Reactions as a Chance for New Tubulin Binders.
Chemistry, 28, 2022
|
|
