5LVX
 
 | | Crystal structure of glucocerebrosidase with an inhibitory quinazoline modulator | | Descriptor: | 11-[(2~{R})-2-[(2-pyridin-3-ylquinazolin-4-yl)amino]-2,3-dihydro-1~{H}-inden-5-yl]undec-10-ynoic acid, 11-[(2~{S})-2-[(2-pyridin-3-ylquinazolin-4-yl)amino]-2,3-dihydro-1~{H}-inden-5-yl]undec-10-ynoic acid, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Zheng, J, Chen, L, Skinner, O.S, Lansbury, P, Skerlj, R, Mrosek, M, Heunisch, U, Krapp, S, Weigand, S, Charrow, J, Schwake, M, Kelleher, N.L, Silverman, R.B, Krainc, D. | | Deposit date: | 2016-09-14 | | Release date: | 2017-10-25 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | beta-Glucocerebrosidase Modulators Promote Dimerization of beta-Glucocerebrosidase and Reveal an Allosteric Binding Site.
J. Am. Chem. Soc., 140, 2018
|
|
5LPL
 
 | | Crystal structure of the bromodomain of human CREBBP bound to the inhibitor XDM3c | | Descriptor: | CREB-binding protein, ~{N}-[(1~{R},2~{R})-7-chloranyl-2-oxidanyl-1,2,3,4-tetrahydronaphthalen-1-yl]-4-ethanoyl-3-ethyl-5-methyl-1~{H}-pyrrole-2-carboxamide | | Authors: | Wohlwend, D, Huegle, M. | | Deposit date: | 2016-08-13 | | Release date: | 2017-08-16 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Beyond the BET Family: Targeting CBP/p300 with 4-Acyl Pyrroles.
Angew. Chem. Int. Ed. Engl., 56, 2017
|
|
6GJ5
 
 | | CRYSTAL STRUCTURE OF KRAS G12D (GPPCP) IN COMPLEX WITH 15 | | Descriptor: | (3~{S})-3-[2-[(2~{R})-pyrrolidin-2-yl]-1~{H}-indol-3-yl]-2,3-dihydroisoindol-1-one, GTPase KRas, MAGNESIUM ION, ... | | Authors: | Kessler, D, Mcconnell, D.M, Mantoulidis, A. | | Deposit date: | 2018-05-16 | | Release date: | 2019-07-31 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.499 Å) | | Cite: | Drugging an undruggable pocket on KRAS.
Proc.Natl.Acad.Sci.USA, 116, 2019
|
|
3CLT
 
 | | Crystal structure of the R236E mutant of Methylophilus methylotrophus ETF | | Descriptor: | ADENOSINE MONOPHOSPHATE, Electron transfer flavoprotein subunit alpha, Electron transfer flavoprotein subunit beta, ... | | Authors: | Katona, G, Leys, D. | | Deposit date: | 2008-03-20 | | Release date: | 2008-04-08 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Probing the dynamic interface between trimethylamine dehydrogenase (TMADH) and electron transferring flavoprotein (ETF) in the TMADH-2ETF complex: role of the Arg-alpha237 (ETF) and Tyr-442 (TMADH) residue pair.
Biochemistry, 47, 2008
|
|
5LPM
 
 | | Crystal structure of the bromodomain of human Ep300 bound to the inhibitor XDM3d | | Descriptor: | ACETATE ION, Histone acetyltransferase p300, ~{N}-[(1~{S},2~{S})-7-chloranyl-2-oxidanyl-1,2,3,4-tetrahydronaphthalen-1-yl]-4-ethanoyl-3-ethyl-5-methyl-1~{H}-pyrrole -2-carboxamide | | Authors: | Huegle, M, Wohlwend, D, Gerhardt, S. | | Deposit date: | 2016-08-13 | | Release date: | 2017-08-16 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Beyond the BET Family: Targeting CBP/p300 with 4-Acyl Pyrroles.
Angew. Chem. Int. Ed. Engl., 56, 2017
|
|
5LMD
 
 | | The crystal structure of hCA II in complex with a benzoxaborole inhibitor | | Descriptor: | 1-[7,7-bis(oxidanyl)-8-oxa-7-boranuidabicyclo[4.3.0]nona-1,3,5-trien-4-yl]-3-(2-methoxy-5-methyl-phenyl)urea, Carbonic anhydrase 2, ZINC ION | | Authors: | De Simone, G, Alterio, V, Esposito, D, Di Fiore, A. | | Deposit date: | 2016-07-29 | | Release date: | 2016-10-19 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Benzoxaborole as a new chemotype for carbonic anhydrase inhibition.
Chem.Commun.(Camb.), 52, 2016
|
|
5LOC
 
 | |
5LCN
 
 | |
3M3X
 
 | | Crystal structure of human carbonic anhydrase isozyme II with 4-{2-[N-(6-methoxy-5-nitropyrimidin-4-yl)amino]ethyl}benzenesulfonamide | | Descriptor: | 4-{2-[(6-methoxy-5-nitropyrimidin-4-yl)amino]ethyl}benzenesulfonamide, Carbonic anhydrase 2, DIMETHYL SULFOXIDE, ... | | Authors: | Grazulis, S, Manakova, E, Golovenko, D. | | Deposit date: | 2010-03-10 | | Release date: | 2010-10-20 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.68 Å) | | Cite: | 4-[N-(Substituted 4-pyrimidinyl)amino]benzenesulfonamides as inhibitors of carbonic anhydrase isozymes I, II, VII, and XIII
Bioorg.Med.Chem., 18, 2010
|
|
3M4J
 
 | | Crystal structure of N-acetyl-L-ornithine transcarbamylase complexed with PALAO | | Descriptor: | N-acetylornithine carbamoyltransferase, N~2~-acetyl-N~5~-(phosphonoacetyl)-L-ornithine, SULFATE ION | | Authors: | Li, Y, Yu, X, Allewell, N.M, Tuchman, M, Shi, D. | | Deposit date: | 2010-03-11 | | Release date: | 2010-07-28 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Reversible Post-Translational Carboxylation Modulates the Enzymatic Activity of N-Acetyl-l-ornithine Transcarbamylase.
Biochemistry, 49, 2010
|
|
3M5E
 
 | | Crystal structure of human carbonic anhydrase isozyme II with 4-{[N-(6-chloro-5-formyl-2-methylthiopyrimidin-4-yl)amino]methyl}benzenesulfonamide | | Descriptor: | 4-({[6-chloro-5-formyl-2-(methylsulfanyl)pyrimidin-4-yl]amino}methyl)benzenesulfonamide, Carbonic anhydrase 2, DIMETHYL SULFOXIDE, ... | | Authors: | Grazulis, S, Manakova, E, Golovenko, D. | | Deposit date: | 2010-03-12 | | Release date: | 2010-10-20 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | 4-[N-(Substituted 4-pyrimidinyl)amino]benzenesulfonamides as inhibitors of carbonic anhydrase isozymes I, II, VII, and XIII
Bioorg.Med.Chem., 18, 2010
|
|
3M98
 
 | |
5LJN
 
 | | Structure of the HOIP PUB domain bound to SPATA2 PIM peptide | | Descriptor: | E3 ubiquitin-protein ligase RNF31, GLYCEROL, SULFATE ION, ... | | Authors: | Elliott, P.R, Komander, D. | | Deposit date: | 2016-07-18 | | Release date: | 2016-08-24 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.701 Å) | | Cite: | SPATA2 Links CYLD to LUBAC, Activates CYLD, and Controls LUBAC Signaling.
Mol.Cell, 63, 2016
|
|
3CLS
 
 | | Crystal structure of the R236C mutant of ETF from Methylophilus methylotrophus | | Descriptor: | ADENOSINE MONOPHOSPHATE, Electron transfer flavoprotein subunit alpha, Electron transfer flavoprotein subunit beta, ... | | Authors: | Katona, G, Leys, D. | | Deposit date: | 2008-03-20 | | Release date: | 2008-04-08 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Probing the dynamic interface between trimethylamine dehydrogenase (TMADH) and electron transferring flavoprotein (ETF) in the TMADH-2ETF complex: role of the Arg-alpha237 (ETF) and Tyr-442 (TMADH) residue pair.
Biochemistry, 47, 2008
|
|
5LOA
 
 | |
6GUS
 
 | |
6GJ7
 
 | | CRYSTAL STRUCTURE OF KRAS G12D (GPPCP) IN COMPLEX WITH 22 | | Descriptor: | (3~{S})-5-oxidanyl-3-[2-[[[1-(phenylmethyl)indol-6-yl]methylamino]methyl]-1~{H}-indol-3-yl]-2,3-dihydroisoindol-1-one, GTPase KRas, MAGNESIUM ION, ... | | Authors: | Kessler, D, Mcconnell, D.M, Mantoulidis, A. | | Deposit date: | 2018-05-16 | | Release date: | 2019-07-31 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.67 Å) | | Cite: | Drugging an undruggable pocket on KRAS.
Proc.Natl.Acad.Sci.USA, 116, 2019
|
|
6TCK
 
 | | Crystal structure of the ATP binding domain of S. aureus GyrB complexed with ULD-2 | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 2-[[3,4-bis(chloranyl)-5-methyl-1~{H}-pyrrol-2-yl]carbonylamino]-4-phenylmethoxy-1,3-benzothiazole-6-carboxylic acid, CALCIUM ION, ... | | Authors: | Welin, M, Kimbung, R, Focht, D. | | Deposit date: | 2019-11-06 | | Release date: | 2020-09-23 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Rational design of balanced dual-targeting antibiotics with limited resistance.
Plos Biol., 18, 2020
|
|
4UED
 
 | |
6TU9
 
 | | The ROR1 Pseudokinase Domain Bound To Ponatinib | | Descriptor: | 3-(imidazo[1,2-b]pyridazin-3-ylethynyl)-4-methyl-N-{4-[(4-methylpiperazin-1-yl)methyl]-3-(trifluoromethyl)phenyl}benzam ide, Inactive tyrosine-protein kinase transmembrane receptor ROR1 | | Authors: | Mathea, S, Preuss, F, Chatterjee, D, Niininen, W, Ungureanu, D, Shin, D, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Knapp, S. | | Deposit date: | 2020-01-04 | | Release date: | 2020-01-22 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.94 Å) | | Cite: | Structural Insights into Pseudokinase Domains of Receptor Tyrosine Kinases.
Mol.Cell, 79, 2020
|
|
8J03
 
 | |
6TX4
 
 | |
6TX9
 
 | |
6UD2
 
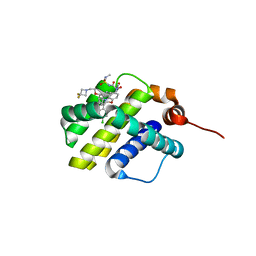 | | co-crystal structure of compound 1 bound to human Mcl-1 | | Descriptor: | (4S,7aR,9aR,10S,11E,18R)-6'-chloro-10-[2-(3,3-difluoroazetidin-1-yl)ethoxy]-N-(dimethylsulfamoyl)-18-hydroxy-15-methyl-16-oxo-3',4',7,7a,8,9,9a,10,13,14,15,16,17,18-tetradecahydro-2'H,3H,5H-spiro[1,19-(ethanediylidene)cyclobuta[n][1,4]oxazepino[4,3-a][1,8]diazacyclohexadecine-4,1'-naphthalene]-18-carboxamide, Induced myeloid leukemia cell differentiation protein Mcl-1 | | Authors: | Huang, X, Whittington, D. | | Deposit date: | 2019-09-18 | | Release date: | 2019-12-04 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Discovery and in Vivo Evaluation of Macrocyclic Mcl-1 Inhibitors Featuring an alpha-Hydroxy Phenylacetic Acid Pharmacophore or Bioisostere.
J.Med.Chem., 62, 2019
|
|
6U1S
 
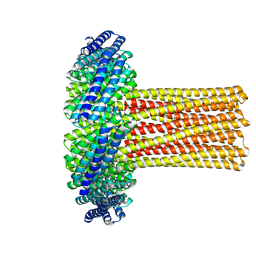 | | Cryo-EM structure of a de novo designed 16-helix transmembrane nanopore, TMHC8_R. | | Descriptor: | de novo designed 16-helix transmembrane nanopore, TMHC8_R | | Authors: | Johnson, M.J, Reggiano, G, Xu, C, Lu, P, Hsia, Y, Brunette, T.J, DiMaio, F, Baker, D, Kollman, J. | | Deposit date: | 2019-08-16 | | Release date: | 2020-08-19 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (7.6 Å) | | Cite: | Computational Design of Transmembrane Channels
To Be Published
|
|
