4N1E
 
 | | Structural evidence for antigen receptor evolution | | Descriptor: | Lysozyme C, immunoglobulin variable light chain domain | | Authors: | Langley, D.B, Rouet, R, Stock, D, Christ, D. | | Deposit date: | 2013-10-04 | | Release date: | 2014-10-29 | | Last modified: | 2018-04-18 | | Method: | X-RAY DIFFRACTION (2.23 Å) | | Cite: | Structural reconstruction of protein ancestry.
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
6ZKA
 
 | | Membrane domain of open complex I during turnover | | Descriptor: | 1,2-DIACYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 1,2-DIACYL-SN-GLYCERO-3-PHOSPHOETHANOLAMINE, 2-decyl-5,6-dimethoxy-3-methylcyclohexa-2,5-diene-1,4-dione, ... | | Authors: | Kampjut, D, Sazanov, L.A. | | Deposit date: | 2020-06-30 | | Release date: | 2020-10-07 | | Last modified: | 2020-11-11 | | Method: | ELECTRON MICROSCOPY (2.5 Å) | | Cite: | The coupling mechanism of mammalian respiratory complex I.
Science, 370, 2020
|
|
6VU5
 
 | | Structure of G-alpha-q bound to its chaperone Ric-8A | | Descriptor: | Guanine nucleotide-binding protein G(q) subunit alpha, Resistance to inhibitors of cholinesterase-8A (Ric-8A) | | Authors: | Seven, A.B, Hilger, D. | | Deposit date: | 2020-02-14 | | Release date: | 2020-03-18 | | Last modified: | 2020-03-25 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Structures of G alpha Proteins in Complex with Their Chaperone Reveal Quality Control Mechanisms.
Cell Rep, 30, 2020
|
|
6N52
 
 | | Metabotropic Glutamate Receptor 5 Apo Form | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Metabotropic glutamate receptor 5 | | Authors: | Koehl, A, Hu, H, Feng, D, Sun, B, Weis, W.I, Skiniotis, G.S, Mathiesen, J.M, Kobilka, B.K. | | Deposit date: | 2018-11-20 | | Release date: | 2019-01-23 | | Last modified: | 2020-07-29 | | Method: | ELECTRON MICROSCOPY (4 Å) | | Cite: | Structural insights into the activation of metabotropic glutamate receptors.
Nature, 566, 2019
|
|
6XKW
 
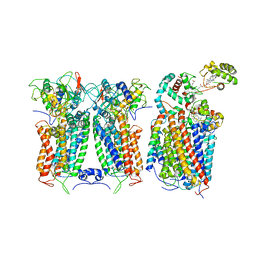 | | R. capsulatus CIII2CIV bipartite super-complex (SC-2A) with CcoH/cy | | Descriptor: | COPPER (II) ION, Cbb3-type cytochrome c oxidase subunit CcoP, Cytochrome b, ... | | Authors: | Steimle, S, Van Eeuwen, T, Ozturk, Y, Kim, H.J, Braitbard, M, Selamoglu, N, Garcia, B.A, Schneidman-Duhovny, D, Murakami, K, Daldal, F. | | Deposit date: | 2020-06-27 | | Release date: | 2020-12-30 | | Last modified: | 2021-03-03 | | Method: | ELECTRON MICROSCOPY (5.2 Å) | | Cite: | Cryo-EM structures of engineered active bc 1 -cbb 3 type CIII 2 CIV super-complexes and electronic communication between the complexes.
Nat Commun, 12, 2021
|
|
6XLV
 
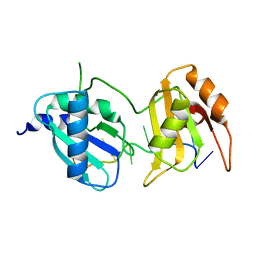 | |
6N51
 
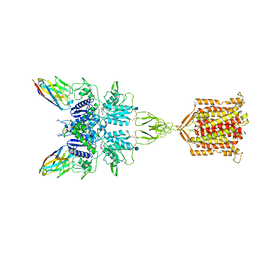 | | Metabotropic Glutamate Receptor 5 bound to L-quisqualate and Nb43 | | Descriptor: | (S)-2-AMINO-3-(3,5-DIOXO-[1,2,4]OXADIAZOLIDIN-2-YL)-PROPIONIC ACID, 2-acetamido-2-deoxy-beta-D-glucopyranose, Metabotropic glutamate receptor 5, ... | | Authors: | Koehl, A, Hu, H, Feng, D, Sun, B, Weis, W.I, Skiniotis, G.S, Mathiesen, J.M, Kobilka, B.K. | | Deposit date: | 2018-11-20 | | Release date: | 2019-01-23 | | Last modified: | 2020-07-29 | | Method: | ELECTRON MICROSCOPY (4 Å) | | Cite: | Structural insights into the activation of metabotropic glutamate receptors.
Nature, 566, 2019
|
|
6VOL
 
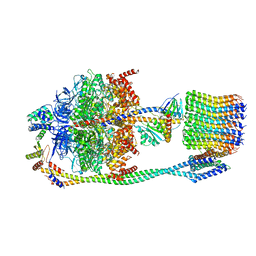 | | Chloroplast ATP synthase (R2, CF1FO) | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, ATP synthase delta chain, ... | | Authors: | Yang, J.-H, Williams, D, Kandiah, E, Fromme, P, Chiu, P.-L. | | Deposit date: | 2020-01-30 | | Release date: | 2020-09-09 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (4.06 Å) | | Cite: | Structural basis of redox modulation on chloroplast ATP synthase.
Commun Biol, 3, 2020
|
|
6N86
 
 | |
5D9C
 
 | | Luciferin-regenerating enzyme solved by SIRAS using XFEL (refined against Hg derivative data) | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, Luciferin regenerating enzyme, MAGNESIUM ION, ... | | Authors: | Yamashita, K, Pan, D, Okuda, T, Murai, T, Kodan, A, Yamaguchi, T, Gomi, K, Kajiyama, N, Kato, H, Ago, H, Yamamoto, M, Nakatsu, T. | | Deposit date: | 2015-08-18 | | Release date: | 2015-09-23 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | An isomorphous replacement method for efficient de novo phasing for serial femtosecond crystallography.
Sci Rep, 5, 2015
|
|
6XKX
 
 | | R. capsulatus CIII2CIV tripartite super-complex, conformation A (SC-1A) | | Descriptor: | COPPER (II) ION, Cbb3-type cytochrome c oxidase subunit CcoP,Cytochrome c-type cyt cy, Cytochrome b, ... | | Authors: | Steimle, S, Van Eeuwen, T, Ozturk, Y, Kim, H.J, Braitbard, M, Selamoglu, N, Garcia, B.A, Schneidman-Duhovny, D, Murakami, K, Daldal, F. | | Deposit date: | 2020-06-27 | | Release date: | 2020-12-30 | | Last modified: | 2021-03-03 | | Method: | ELECTRON MICROSCOPY (6.1 Å) | | Cite: | Cryo-EM structures of engineered active bc 1 -cbb 3 type CIII 2 CIV super-complexes and electronic communication between the complexes.
Nat Commun, 12, 2021
|
|
6XLQ
 
 | | Crystal Structure of the Human BTN3A1 Ectodomain in Complex with the CTX-2026 Fab | | Descriptor: | Butyrophilin subfamily 3 member A1, CTX-2026 Heavy Chain, CTX-2026 Light Chain | | Authors: | Payne, K.K, Mine, J.A, Biswas, S, Chaurio, R.A, Perales-Puchalt, A, Anadon, C.M, Costich, T.L, Harro, C.M, Walrath, J, Ming, Q, Tcyganov, E, Buras, A.L, Rigolizzo, K.E, Mandal, G, Lajoie, J, Ophir, M, Tchou, J, Marchion, D, Luca, V.C, Bobrowicz, P, McLaughlin, B, Eskiocak, U, Schmidt, M, Cubillos-Ruiz, J.R, Rodriguez, P.C, Gabrilovich, D.I, Conejo-Garcia, J.R. | | Deposit date: | 2020-06-29 | | Release date: | 2020-09-02 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | BTN3A1 governs antitumor responses by coordinating alpha beta and gamma delta T cells.
Science, 369, 2020
|
|
5D70
 
 | | Crystal structure of MOR03929, a neutralizing anti-human GM-CSF antibody Fab fragment in complex with human GM-CSF | | Descriptor: | Granulocyte-macrophage colony-stimulating factor, Immunglobulin G1 Fab fragment, heavy chain, ... | | Authors: | Eylenstein, R, Weinfurtner, D, Steidl, S, Boettcher, J, Augustin, M. | | Deposit date: | 2015-08-13 | | Release date: | 2015-10-14 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.06 Å) | | Cite: | Molecular basis of in vitro affinity maturation and functional evolution of a neutralizing anti-human GM-CSF antibody.
Mabs, 8, 2016
|
|
5DAZ
 
 | |
6VRM
 
 | | T cell receptor-p53-HLA-A2 complex | | Descriptor: | Beta-2-microglobulin, Cellular tumor antigen p53 peptide, MHC class I antigen, ... | | Authors: | Wu, D, Gallagher, D.T, Gowthaman, R, Pierce, B.G, Mariuzza, R.A. | | Deposit date: | 2020-02-08 | | Release date: | 2020-06-17 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.61 Å) | | Cite: | Structural basis for oligoclonal T cell recognition of a shared p53 cancer neoantigen.
Nat Commun, 11, 2020
|
|
6W6K
 
 | | 30S-Activated-high-Mg2+ | | Descriptor: | 16S rRNA, 30S ribosomal protein S10, 30S ribosomal protein S11, ... | | Authors: | Jahagirdar, D, Jha, V, Basu, B, Gomez-Blanco, J, Vargas, J, Ortega, J. | | Deposit date: | 2020-03-17 | | Release date: | 2020-10-21 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Alternative conformations and motions adopted by 30S ribosomal subunits visualized by cryo-electron microscopy.
Rna, 26, 2020
|
|
5D7S
 
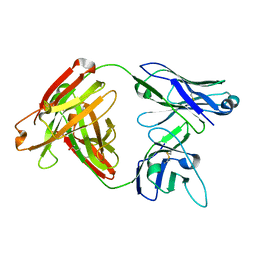 | | Crystal structure of MOR04357, a neutralizing anti-human GM-CSF antibody Fab fragment | | Descriptor: | (2S,3S)-butane-2,3-diol, Immunglobulin G1 Fab fragment, heavy chain, ... | | Authors: | Eylenstein, R, Weinfurtner, D, Steidl, S, Boettcher, J, Augustin, M. | | Deposit date: | 2015-08-14 | | Release date: | 2015-10-14 | | Last modified: | 2016-01-13 | | Method: | X-RAY DIFFRACTION (1.88 Å) | | Cite: | Molecular basis of in vitro affinity maturation and functional evolution of a neutralizing anti-human GM-CSF antibody.
Mabs, 8, 2016
|
|
6XOX
 
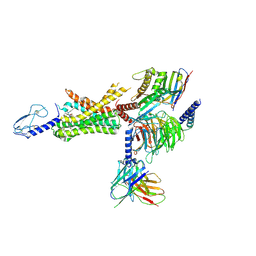 | | cryo-EM of human GLP-1R bound to non-peptide agonist LY3502970 | | Descriptor: | 3-[(1S,2S)-1-(5-[(4S)-2,2-dimethyloxan-4-yl]-2-{(4S)-2-(4-fluoro-3,5-dimethylphenyl)-3-[3-(4-fluoro-1-methyl-1H-indazol-5-yl)-2-oxo-2,3-dihydro-1H-imidazol-1-yl]-4-methyl-2,4,6,7-tetrahydro-5H-pyrazolo[4,3-c]pyridine-5-carbonyl}-1H-indol-1-yl)-2-methylcyclopropyl]-1,2,4-oxadiazol-5(4H)-one, Alpha subunit of Gs with N-terminus swapped with equivalent residues in Gi,Guanine nucleotide-binding protein G(s) subunit alpha isoforms XLas, Glucagon-like peptide 1 receptor, ... | | Authors: | Sun, B, Kobilka, B.K, Sloop, K.W, Feng, D, Kobilka, T.S. | | Deposit date: | 2020-07-07 | | Release date: | 2020-11-18 | | Last modified: | 2020-12-09 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Structural basis for GLP-1 receptor activation by LY3502970, an orally active nonpeptide agonist.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
4LJC
 
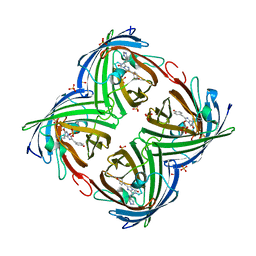 | | Structure of an X-ray-induced photobleached state of IrisFP | | Descriptor: | Green to red photoconvertible GPF-like protein EosFP, SULFATE ION, SULFITE ION | | Authors: | Duan, C, Adam, V, Byrdin, M, Ridard, J, Kieffer-Jacquinod, S, Morlot, C, Arcizet, D, Demachy, I, Bourgeois, D. | | Deposit date: | 2013-07-04 | | Release date: | 2013-10-09 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.86 Å) | | Cite: | Structural evidence for a two-regime photobleaching mechanism in a reversibly switchable fluorescent protein.
J.Am.Chem.Soc., 135, 2013
|
|
5CSU
 
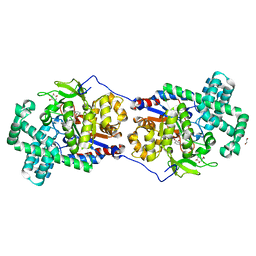 | | Disproportionating enzyme 1 from Arabidopsis - acarviostatin soak | | Descriptor: | 1,2-ETHANEDIOL, 4-alpha-glucanotransferase DPE1, chloroplastic/amyloplastic, ... | | Authors: | O'Neill, E.C, Stevenson, C.E.M, Tantanarat, K, Latousakis, D, Donaldson, M.I, Rejzek, M, Limpaseni, T, Smith, A.M, Field, R.A, Lawson, D.M. | | Deposit date: | 2015-07-23 | | Release date: | 2015-11-04 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.53 Å) | | Cite: | Structural Dissection of the Maltodextrin Disproportionation Cycle of the Arabidopsis Plastidial Disproportionating Enzyme 1 (DPE1).
J.Biol.Chem., 290, 2015
|
|
6NES
 
 | | FAD-dependent monooxygenase TropB from T. stipitatus | | Descriptor: | CHLORIDE ION, FAD-dependent monooxygenase tropB, FLAVIN-ADENINE DINUCLEOTIDE, ... | | Authors: | Rodriguez Benitez, A, Tweedy, S.E, Baker Dockrey, S.A, Lukowski, A.L, Wymore, T, Khare, D, Brooks, C.L, Palfey, B.A, Smith, J.L, Narayan, A.R.H. | | Deposit date: | 2018-12-18 | | Release date: | 2019-08-14 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Structural basis for selectivity in flavin-dependent monooxygenase-catalyzed oxidative dearomatization.
Acs Catalysis, 9, 2019
|
|
6XUZ
 
 | | CRYSTAL STRUCTURE OF BRD4-BD1 WITH COMPOUND 4 | | Descriptor: | 6-[1-[(2~{S})-1-methoxypropan-2-yl]-6-[(3~{S})-3-methylmorpholin-4-yl]imidazo[4,5-c]pyridin-2-yl]-3-methyl-~{N}-propan-2-yl-[1,2,4]triazolo[4,3-a]pyrazin-8-amine, Bromodomain-containing protein 4 | | Authors: | Bader, G, Kessler, D, Wolkerstorfer, B. | | Deposit date: | 2020-01-21 | | Release date: | 2020-07-08 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.07 Å) | | Cite: | PI by NMR: Probing CH-pi Interactions in Protein-Ligand Complexes by NMR Spectroscopy.
Angew.Chem.Int.Ed.Engl., 59, 2020
|
|
6XV3
 
 | | CRYSTAL STRUCTURE OF BRD4-BD1 WITH COMPOUND 3 | | Descriptor: | 3-methyl-6-[6-[(3~{S})-3-methylmorpholin-4-yl]-1-[(1~{S})-1-phenylethyl]imidazo[4,5-c]pyridin-2-yl]-~{N}-propan-2-yl-[1,2,4]triazolo[4,3-a]pyrazin-8-amine, Bromodomain-containing protein 4 | | Authors: | Bader, G, Kessler, D, Wolkerstorfer, B. | | Deposit date: | 2020-01-21 | | Release date: | 2020-07-08 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.47 Å) | | Cite: | PI by NMR: Probing CH-pi Interactions in Protein-Ligand Complexes by NMR Spectroscopy.
Angew.Chem.Int.Ed.Engl., 59, 2020
|
|
6VOK
 
 | | Chloroplast ATP synthase (R3, CF1) | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, ATP synthase delta chain, ... | | Authors: | Yang, J.-H, Williams, D, Kandiah, E, Fromme, P, Chiu, P.-L. | | Deposit date: | 2020-01-30 | | Release date: | 2020-09-09 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (3.85 Å) | | Cite: | Structural basis of redox modulation on chloroplast ATP synthase.
Commun Biol, 3, 2020
|
|
1MI0
 
 | | Crystal Structure of the redesigned protein G variant NuG2 | | Descriptor: | immunoglobulin-binding protein G | | Authors: | Nauli, S, Kuhlman, B, Le Trong, I, Stenkamp, R.E, Teller, D.C, Baker, D. | | Deposit date: | 2002-08-21 | | Release date: | 2002-09-18 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Crystal structures and increased stabilization of the protein G variants with switched folding pathways NuG1 and NuG2
Biochemistry, 11, 2002
|
|
