4M6I
 
 | | Structure of the reduced, Zn-bound form of Mycobacterium tuberculosis peptidoglycan amidase Rv3717 | | Descriptor: | Peptidoglycan Amidase Rv3717, ZINC ION | | Authors: | Prigozhin, D.M, Mavrici, D, Huizar, J.P, Vansell, H.J, Alber, T, TB Structural Genomics Consortium (TBSGC) | | Deposit date: | 2013-08-09 | | Release date: | 2013-09-18 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.666 Å) | | Cite: | Structural and Biochemical Analyses of Mycobacterium tuberculosis N-Acetylmuramyl-L-alanine Amidase Rv3717 Point to a Role in Peptidoglycan Fragment Recycling.
J.Biol.Chem., 288, 2013
|
|
4M1J
 
 | | Crystal structure of Pseudomonas aeruginosa PvdQ in complex with a transition state analogue | | Descriptor: | Acyl-homoserine lactone acylase PvdQ subunit alpha, Acyl-homoserine lactone acylase PvdQ subunit beta, GLYCEROL, ... | | Authors: | Wu, R, Clevenger, K, Er, J, Fast, W.L, Liu, D. | | Deposit date: | 2013-08-02 | | Release date: | 2013-08-28 | | Last modified: | 2017-11-15 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Rational Design of a Transition State Analogue with Picomolar Affinity for Pseudomonas aeruginosa PvdQ, a Siderophore Biosynthetic Enzyme.
Acs Chem.Biol., 8, 2013
|
|
4M4X
 
 | |
4LX5
 
 | | X-ray crystal structure of the M6" riboswitch aptamer bound to pyrimido[4,5-d]pyrimidine-2,4-diamine (PPDA) | | Descriptor: | MAGNESIUM ION, Mutated adenine riboswitch aptamer, pyrimido[4,5-d]pyrimidine-2,4-diamine | | Authors: | Dunstan, M.S, Leys, D. | | Deposit date: | 2013-07-29 | | Release date: | 2014-07-16 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.13 Å) | | Cite: | Modular riboswitch toolsets for synthetic genetic control in diverse bacterial species.
J.Am.Chem.Soc., 136, 2014
|
|
4M6H
 
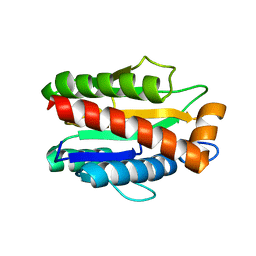 | | Structure of the reduced, metal-free form of Mycobacterium tuberculosis peptidoglycan amidase Rv3717 | | Descriptor: | Peptidoglycan Amidase Rv3717 | | Authors: | Prigozhin, D.M, Mavrici, D, Huizar, J.P, Vansell, H.J, Alber, T, TB Structural Genomics Consortium (TBSGC) | | Deposit date: | 2013-08-09 | | Release date: | 2013-09-18 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.194 Å) | | Cite: | Structural and Biochemical Analyses of Mycobacterium tuberculosis N-Acetylmuramyl-L-alanine Amidase Rv3717 Point to a Role in Peptidoglycan Fragment Recycling.
J.Biol.Chem., 288, 2013
|
|
4M4P
 
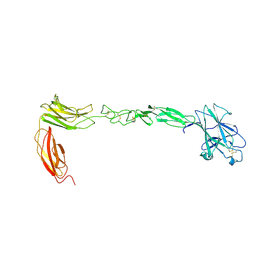 | | Crystal structure of EPHA4 ectodomain | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Ephrin type-A receptor 4 | | Authors: | Xu, K, Tsvetkova-Robev, D, Xu, Y, Goldgur, Y, Chan, Y.-P, Himanen, J.P, Nikolov, D.B. | | Deposit date: | 2013-08-07 | | Release date: | 2013-10-30 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.081 Å) | | Cite: | Insights into Eph receptor tyrosine kinase activation from crystal structures of the EphA4 ectodomain and its complex with ephrin-A5.
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
4DLI
 
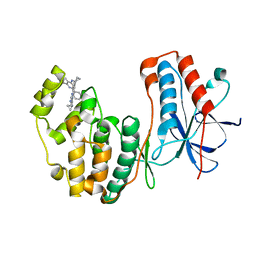 | | Human p38 MAP kinase in complex with RL87 | | Descriptor: | Mitogen-activated protein kinase 14, N~4~-cyclopropyl-2-phenylquinazoline-4,7-diamine | | Authors: | Gruetter, C, Getlik, M, Simard, J.R, Rauh, D. | | Deposit date: | 2012-02-06 | | Release date: | 2013-01-23 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.91 Å) | | Cite: | Fluorophore labeled kinase detects ligands that bind within the MAPK insert of p38alpha kinase.
Plos One, 7, 2012
|
|
3KOY
 
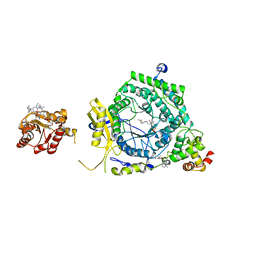 | | Crystal Structure of ornithine 4,5 aminomutase in complex with ornithine (Aerobic) | | Descriptor: | (E)-N~5~-({3-hydroxy-2-methyl-5-[(phosphonooxy)methyl]pyridin-4-yl}methylidene)-L-ornithine, 5'-DEOXYADENOSINE, COBALAMIN, ... | | Authors: | Wolthers, K.R, Levy, C.W, Scrutton, N.S, Leys, D. | | Deposit date: | 2009-11-14 | | Release date: | 2010-01-26 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Large-scale domain dynamics and adenosylcobalamin reorientation orchestrate radical catalysis in ornithine 4,5-aminomutase.
J.Biol.Chem., 285, 2010
|
|
4DM9
 
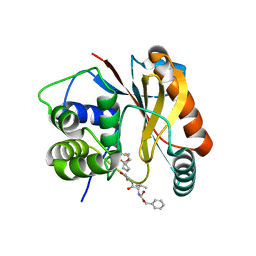 | | The Crystal Structure of Ubiquitin Carboxy-terminal hydrolase L1 (UCHL1) bound to a tripeptide fluoromethyl ketone Z-VAE(OMe)-FMK | | Descriptor: | Tripeptide fluoromethyl ketone inhibitor Z-VAE(OMe)-FMK, Ubiquitin carboxyl-terminal hydrolase isozyme L1 | | Authors: | Davies, C.W, Chaney, J, Korbel, G, Ringe, D, Petsko, G.A, Ploegh, H, Das, C. | | Deposit date: | 2012-02-07 | | Release date: | 2012-05-23 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | The co-crystal structure of ubiquitin carboxy-terminal hydrolase L1 (UCHL1) with a tripeptide fluoromethyl ketone (Z-VAE(OMe)-FMK).
Bioorg.Med.Chem.Lett., 22, 2012
|
|
4DG5
 
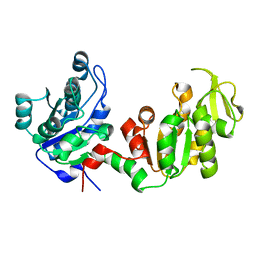 | |
3KP8
 
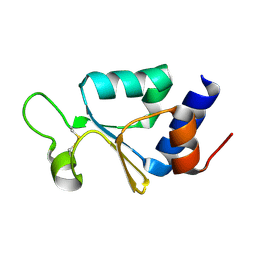 | | The thioredoxin-like domain of a VKOR homolog from Synechococcus sp. | | Descriptor: | VKORC1/thioredoxin domain protein | | Authors: | Li, W, Schulman, S, Dutton, R.J, Boyd, D, Beckwith, J, Rapoport, T.A. | | Deposit date: | 2009-11-15 | | Release date: | 2010-03-02 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.66 Å) | | Cite: | Structure of a bacterial homologue of vitamin K epoxide reductase.
Nature, 463, 2010
|
|
3KPE
 
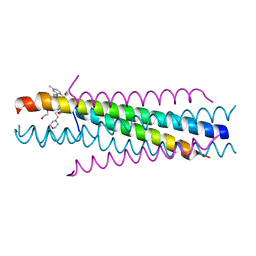 | | Solution structure of the respiratory syncytial virus (RSV)six-helix bundle complexed with TMC353121, a small-moleucule inhibitor of RSV | | Descriptor: | 2-[[6-[[[2-(3-hydroxypropyl)-5-methylphenyl]amino]methyl]-2-[[3-(4-morpholinyl)propyl]amino]-1H-benzimidazol-1-yl]methyl]-6-methyl-3-pyridinol, Fusion glycoprotein F0, TETRAETHYLENE GLYCOL | | Authors: | Roymans, D, De Bondt, H, Arnoult, E, Cummings, M.D, Van Vlijmen, H, Andries, K. | | Deposit date: | 2009-11-16 | | Release date: | 2009-12-22 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.47 Å) | | Cite: | Binding of a potent small-molecule inhibitor of six-helix bundle formation requires interactions with both heptad-repeats of the RSV fusion protein.
Proc.Natl.Acad.Sci.USA, 107, 2010
|
|
3KPL
 
 | | Crystal Structure of HLA B*4402 in complex with EEYLQAFTY a self peptide from the ABCD3 protein | | Descriptor: | Beta-2-microglobulin, EEYLQAFTY, self peptide from the ATP binding cassette protein ABCD3, ... | | Authors: | Macdonald, W.A, Chen, Z, Gras, S, Archbold, J.K, Tynan, F.E, Clements, C.S, Bharadwaj, M, Kjer-Nielsen, L, Saunders, P.M, Wilce, M.C, Crawford, F, Stadinsky, B, Jackson, D, Brooks, A.G, Purcell, A.W, Kappler, J.W, Burrows, S.R, Rossjohn, J, McCluskey, J. | | Deposit date: | 2009-11-16 | | Release date: | 2009-12-22 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.96 Å) | | Cite: | T cell allorecognition via molecular mimicry.
Immunity, 31, 2009
|
|
4DOF
 
 | |
3KPS
 
 | | Crystal Structure of the LC13 TCR in complex with HLA B*4405 bound to EEYLQAFTY a self peptide from the ABCD3 protein | | Descriptor: | Beta-2-microglobulin, EEYLQAFTY, self peptide from the ATP binding cassette protein ABCD3, ... | | Authors: | Macdonald, W.A, Chen, Z, Gras, S, Archbold, J.K, Tynan, F.E, Clements, C.S, Bharadwaj, M, Kjer-Nielsen, L, Saunders, P.M, Wilce, M.C, Crawford, F, Stadinsky, B, Jackson, D, Brooks, A.G, Purcell, A.W, Kappler, J.W, Burrows, S.R, Rossjohn, J, McCluskey, J. | | Deposit date: | 2009-11-16 | | Release date: | 2009-12-22 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | T cell allorecognition via molecular mimicry.
Immunity, 31, 2009
|
|
3KUC
 
 | | Complex of Rap1A(E30D/K31E)GDP with RafRBD(A85K/N71R) | | Descriptor: | CALCIUM ION, GUANOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Filchtinski, D, Sharabi, O, Rueppel, A, Vetter, I.R, Herrmann, C, Shifman, J.M. | | Deposit date: | 2009-11-27 | | Release date: | 2010-03-23 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.92 Å) | | Cite: | What makes Ras an efficient molecular switch: a computational, biophysical, and structural study of Ras-GDP interactions with mutants of Raf.
J.Mol.Biol., 399, 2010
|
|
4DPH
 
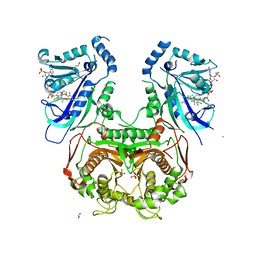 | | Quadruple mutant (N51I+C59R+S108N+I164L) Plasmodium falciparum dihydrofolate reductase-thymidylate synthase (PfDHFR-TS) complexed with P65 and NADPH | | Descriptor: | 2,4-diamino-6-methyl-5-[3-(2,4,5-trichlorophenoxy)propyloxy]pyrimidine, BETA-MERCAPTOETHANOL, Bifunctional dihydrofolate reductase-thymidylate synthase, ... | | Authors: | Yuthavong, Y, Vilaivan, T, Kamchonwongpaisan, S, Charman, S.A, McLennan, D.N, White, K.L, Vivas, L, Bongard, E, Chitnumsub, P, Tarnchompoo, B, Thongphanchang, C, Taweechai, S, Vanichtanakul, J, Arwon, U, Fantauzzi, P, Yuvaniyama, J, Charman, W.N, Matthews, D. | | Deposit date: | 2012-02-13 | | Release date: | 2012-11-14 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.38 Å) | | Cite: | Malarial dihydrofolate reductase as a paradigm for drug development against a resistance-compromised target
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
3KVU
 
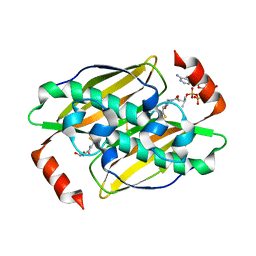 | | Structural basis of the activity and substrate specificity of the fluoroacetyl-CoA FlK - T42S mutant in complex with Acetyl-CoA | | Descriptor: | ACETYL COENZYME *A, Fluoroacetyl-CoA thioesterase FlK | | Authors: | Dias, M.V.B, Huang, F, Chirgadze, D.Y, Tosin, M, Spiteller, D, Valentine, E.F, Leadlay, P.F, Spencer, J.B, Blundell, T.L. | | Deposit date: | 2009-11-30 | | Release date: | 2010-04-28 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural basis for the activity and substrate specificity of fluoroacetyl-CoA thioesterase FlK.
J.Biol.Chem., 285, 2010
|
|
3KW6
 
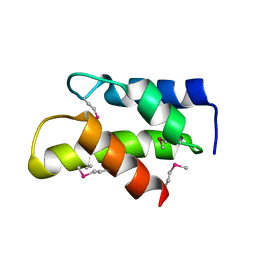 | | Crystal Structure of a domain of 26S proteasome regulatory subunit 8 from homo sapiens. Northeast Structural Genomics Consortium target id HR3102A | | Descriptor: | 26S protease regulatory subunit 8 | | Authors: | Seetharaman, J, Su, M, Wang, D, Janjua, H, Cunningham, K, Owens, L, Xiao, R, Liu, J, Baran, M.C, Acton, T.B, Montelione, G.T, Hunt, J.F, Tong, L, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2009-11-30 | | Release date: | 2009-12-22 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal Structure of a domain of 26S proteasome regulatory subunit 8 from homo sapiens. Northeast Structural Genomics Consortium target id HR3102A
To be Published
|
|
4DJI
 
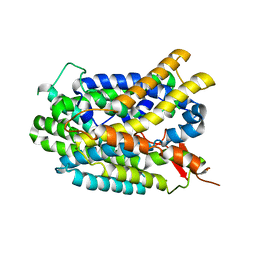 | | Structure of glutamate-GABA antiporter GadC | | Descriptor: | Probable glutamate/gamma-aminobutyrate antiporter | | Authors: | Ma, D, Lu, P.L, Yan, C.Y, Fan, C, Yin, P, Wang, J.W, Shi, Y.G. | | Deposit date: | 2012-02-02 | | Release date: | 2012-03-14 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (3.187 Å) | | Cite: | Structure and mechanism of a glutamate-GABA antiporter
Nature, 483, 2012
|
|
3KRD
 
 | |
4DLQ
 
 | | Crystal structure of the GAIN and HormR domains of CIRL 1/Latrophilin 1 (CL1) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, GLYCEROL, ... | | Authors: | Arac, D, Boucard, A.A, Bolliger, M.F, Nguyen, J, Soltis, M, Sudhof, T.C, Brunger, A.T. | | Deposit date: | 2012-02-06 | | Release date: | 2012-02-22 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | A novel evolutionarily conserved domain of cell-adhesion GPCRs mediates autoproteolysis.
Embo J., 31, 2012
|
|
7XNA
 
 | | Crystal structure of somatostatin receptor 2 (SSTR2) with peptide antagonist CYN 154806 | | Descriptor: | CYN 154806, Somatostatin receptor type 2,Endo-1,4-beta-xylanase | | Authors: | Zhao, W, Han, S, Qiu, N, Feng, W, Lu, M, Yang, D, Wang, M.-W, Wu, B, Zhao, Q. | | Deposit date: | 2022-04-28 | | Release date: | 2022-08-03 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Structural insights into ligand recognition and selectivity of somatostatin receptors.
Cell Res., 32, 2022
|
|
4DQ9
 
 | | Crystal structure of the minor pseudopilin EPSH from the type II secretion system of Vibrio cholerae | | Descriptor: | CHLORIDE ION, General secretion pathway protein H, SODIUM ION | | Authors: | Raghunathan, K, Vago, F.S, Grindem, D, Ball, T, Wedemeyer, W.J, Arvidson, D.N. | | Deposit date: | 2012-02-15 | | Release date: | 2013-03-13 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.59 Å) | | Cite: | The 1.59 angstrom resolution structure of the minor pseudopilin EpsH of Vibrio cholerae reveals a long flexible loop.
Biochim.Biophys.Acta, 1844, 2013
|
|
7XN9
 
 | | Crystal structure of SSTR2 and L-054,522 complex | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, Somatostatin receptor type 2,Endo-1,4-beta-xylanase, tert-butyl (2S)-6-azanyl-2-[[(2R,3S)-3-(1H-indol-3-yl)-2-[[4-(2-oxidanylidene-3H-benzimidazol-1-yl)piperidin-1-yl]carbonylamino]butanoyl]amino]hexanoate | | Authors: | Zhao, W, Han, S, Qiu, N, Feng, W, Lu, M, Yang, D, Wang, M.-W, Wu, B, Zhao, Q. | | Deposit date: | 2022-04-28 | | Release date: | 2022-08-03 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structural insights into ligand recognition and selectivity of somatostatin receptors.
Cell Res., 32, 2022
|
|
