6KJ3
 
 | | 120kV MicroED structure of FUS (37-42) SYSGYS solved from merged datasets at 0.60 A | | Descriptor: | RNA-binding protein FUS | | Authors: | Zhou, H, Luo, F, Luo, Z, Li, D, Liu, C, Li, X. | | Deposit date: | 2019-07-20 | | Release date: | 2019-10-02 | | Last modified: | 2024-03-27 | | Method: | ELECTRON CRYSTALLOGRAPHY (0.6 Å) | | Cite: | Programming Conventional Electron Microscopes for Solving Ultrahigh-Resolution Structures of Small and Macro-Molecules.
Anal.Chem., 91, 2019
|
|
5ZH8
 
 | | Crystal Structure of FmtA from Staphylococcus aureus at 2.58 A | | Descriptor: | GLYCEROL, Protein FmtA | | Authors: | Dalal, V, Kumar, P, Golemi-Kotra, D, Kumar, P. | | Deposit date: | 2018-03-12 | | Release date: | 2019-07-17 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Repurposing an Ancient Protein Core Structure: Structural Studies on FmtA, a Novel Esterase of Staphylococcus aureus.
J.Mol.Biol., 431, 2019
|
|
5YX1
 
 | | Crystal structure of hematopoietic prostaglandin D synthase in complex with U004 | | Descriptor: | GLUTATHIONE, GLYCEROL, Hematopoietic prostaglandin D synthase, ... | | Authors: | Kamo, M, Furubayashi, N, Inaka, K, Aritake, K, Urade, Y, Takaya, D, Tanaka, A. | | Deposit date: | 2017-12-01 | | Release date: | 2018-12-05 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.39 Å) | | Cite: | Crystal structure of hematopoietic prostaglandin D synthase in complex with U004
To Be Published
|
|
1D4A
 
 | | CRYSTAL STRUCTURE OF HUMAN NAD[P]H-QUINONE OXIDOREDUCTASE AT 1.7 A RESOLUTION | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, QUINONE REDUCTASE | | Authors: | Faig, M, Bianchet, M.A, Chen, S, Winski, S, Ross, D, Amzel, L.M. | | Deposit date: | 1999-10-01 | | Release date: | 1999-10-15 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structures of recombinant human and mouse NAD(P)H:quinone oxidoreductases: species comparison and structural changes with substrate binding and release.
Proc.Natl.Acad.Sci.USA, 97, 2000
|
|
1CEZ
 
 | | CRYSTAL STRUCTURE OF A T7 RNA POLYMERASE-T7 PROMOTER COMPLEX | | Descriptor: | DNA (5'-D(P*TP*AP*AP*TP*AP*CP*GP*AP*CP*TP*CP*AP*CP*TP*A)-3'), DNA (5'-D(P*TP*AP*TP*AP*GP*TP*GP*AP*GP*TP*CP*GP*TP*AP*TP*TP*A)-3'), PROTEIN (BACTERIOPHAGE T7 RNA POLYMERASE) | | Authors: | Cheetham, G.M.T, Jeruzalmi, D, Steitz, T.A. | | Deposit date: | 1999-03-11 | | Release date: | 1999-05-21 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural basis for initiation of transcription from an RNA polymerase-promoter complex.
Nature, 399, 1999
|
|
1CLU
 
 | | H-RAS COMPLEXED WITH DIAMINOBENZOPHENONE-BETA,GAMMA-IMIDO-GTP | | Descriptor: | 3-AMINOBENZOPHENONE-4-YL-AMINOHYDROXYPHOSPHINYLAMINOPHOSPHONIC ACID-GUANYLATE ESTER, MAGNESIUM ION, TRANSFORMING PROTEIN P21/H-RAS-1 | | Authors: | Ahmadian, M.R, Zor, T, Vogt, D, Kabsch, W, Selinger, Z, Wittinghofer, A, Scheffzek, K. | | Deposit date: | 1999-05-03 | | Release date: | 1999-05-28 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Guanosine triphosphatase stimulation of oncogenic Ras mutants.
Proc.Natl.Acad.Sci.USA, 96, 1999
|
|
5W9F
 
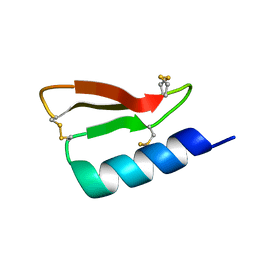 | | Solution structure of the de novo mini protein gHEEE_02 | | Descriptor: | De novo mini protein gHEEE_02 | | Authors: | Pulavarti, S.V.S.R.K, Shaw, E.A, Bahl, C.D, Garry, B.W, Baker, D, Szyperski, T. | | Deposit date: | 2017-06-23 | | Release date: | 2018-07-11 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | Cytosolic expression, solution structures, and molecular dynamics simulation of genetically encodable disulfide-rich de novo designed peptides.
Protein Sci., 27, 2018
|
|
1D2N
 
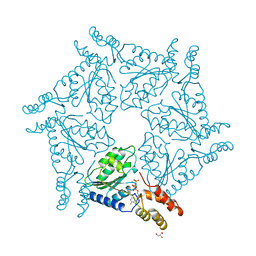 | | D2 DOMAIN OF N-ETHYLMALEIMIDE-SENSITIVE FUSION PROTEIN | | Descriptor: | GLYCEROL, MAGNESIUM ION, N-ETHYLMALEIMIDE-SENSITIVE FUSION PROTEIN, ... | | Authors: | Lenzen, C.U, Steinmann, D, Whiteheart, S.W, Weis, W.I. | | Deposit date: | 1998-06-30 | | Release date: | 1998-10-14 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Crystal structure of the hexamerization domain of N-ethylmaleimide-sensitive fusion protein.
Cell(Cambridge,Mass.), 94, 1998
|
|
2KQE
 
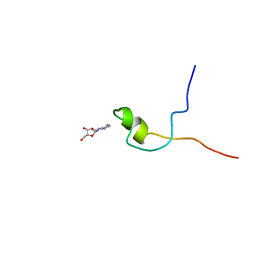 | | Second PBZ domain of human APLF protein in complex with ribofuranosyladenosine | | Descriptor: | ADENOSINE, Aprataxin and PNK-like factor, ZINC ION, ... | | Authors: | Neuhaus, D, Eustermann, S, Brockmann, C, Yang, J. | | Deposit date: | 2009-11-04 | | Release date: | 2010-01-19 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Solution structures of the two PBZ domains from human APLF and their interaction with poly(ADP-ribose).
Nat.Struct.Mol.Biol., 17, 2010
|
|
5XJ7
 
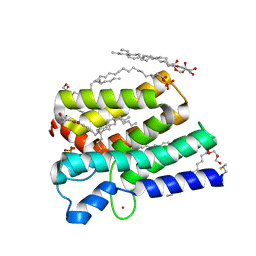 | | Crystal structure of PlsY (YgiH), an integral membrane glycerol 3-phosphate acyltransferase - the acyl phosphate form | | Descriptor: | (2S)-2,3-DIHYDROXYPROPYL(7Z)-PENTADEC-7-ENOATE, Glycerol-3-phosphate acyltransferase, PHOSPHATE ION, ... | | Authors: | Tang, Y, Li, Z, Li, D. | | Deposit date: | 2017-04-30 | | Release date: | 2017-12-06 | | Method: | X-RAY DIFFRACTION (1.772 Å) | | Cite: | Structural insights into the committed step of bacterial phospholipid biosynthesis.
Nat Commun, 8, 2017
|
|
5ZGD
 
 | | hnRNPA1 reversible amyloid core GFGGNDNFG (residues 209-217) determined by X-ray | | Descriptor: | GLY-PHE-GLY-GLY-ASN-ASP-ASN-PHE-GLY | | Authors: | Gui, X, Xie, M, Zhao, M, Luo, F, He, J, Li, D, Liu, C. | | Deposit date: | 2018-03-08 | | Release date: | 2019-04-03 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.401 Å) | | Cite: | Structural basis for reversible amyloids of hnRNPA1 elucidates their role in stress granule assembly.
Nat Commun, 10, 2019
|
|
1CTI
 
 | |
1CUN
 
 | | CRYSTAL STRUCTURE OF REPEATS 16 AND 17 OF CHICKEN BRAIN ALPHA SPECTRIN | | Descriptor: | PROTEIN (ALPHA SPECTRIN) | | Authors: | Grum, V.L, Li, D, MacDonald, R.I, Mondragon, A. | | Deposit date: | 1999-08-20 | | Release date: | 1999-10-06 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structures of two repeats of spectrin suggest models of flexibility.
Cell(Cambridge,Mass.), 98, 1999
|
|
1CZ2
 
 | | SOLUTION STRUCTURE OF WHEAT NS-LTP COMPLEXED WITH PROSTAGLANDIN B2. | | Descriptor: | NONSPECIFIC LIPID TRANSFER PROTEIN, PROSTAGLANDIN B2 | | Authors: | Tassin-Moindrot, S, Caille, A, Douliez, J.-P, Marion, D, Vovelle, F. | | Deposit date: | 1999-09-01 | | Release date: | 2000-09-27 | | Last modified: | 2022-02-16 | | Method: | SOLUTION NMR | | Cite: | The wide binding properties of a wheat nonspecific lipid transfer protein. Solution structure of a complex with prostaglandin B2.
Eur.J.Biochem., 267, 2000
|
|
2KRL
 
 | | The ensemble of the solution global structures of the 102-nt ribosome binding structure element of the turnip crinkle virus 3' UTR RNA | | Descriptor: | RNA (102-MER) | | Authors: | Zuo, X, Wang, J, Yu, P, Eyler, D, Xu, H, Starich, M, Tiede, D, Simon, A, Kasprzak, W, Schwieters, C, Shapiro, B. | | Deposit date: | 2009-12-18 | | Release date: | 2011-02-09 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR, SOLUTION SCATTERING | | Cite: | Solution structure of the cap-independent translational enhancer and ribosome-binding element in the 3' UTR of turnip crinkle virus.
Proc.Natl.Acad.Sci.USA, 107, 2010
|
|
1CJX
 
 | | CRYSTAL STRUCTURE OF PSEUDOMONAS FLUORESCENS HPPD | | Descriptor: | 4-HYDROXYPHENYLPYRUVATE DIOXYGENASE, ACETATE ION, ETHYL MERCURY ION, ... | | Authors: | Serre, L, Sailland, A, Sy, D, Boudec, P, Rolland, A, Pebay-Peroulla, E, Cohen-Addad, C. | | Deposit date: | 1999-04-20 | | Release date: | 2000-04-26 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal structure of Pseudomonas fluorescens 4-hydroxyphenylpyruvate dioxygenase: an enzyme involved in the tyrosine degradation pathway.
Structure Fold.Des., 7, 1999
|
|
5Z1V
 
 | | Crystal structure of AvrPib | | Descriptor: | AvrPib protein | | Authors: | Zhang, X, He, D, Zhao, Y.X, Taylor, I.A, Peng, Y.L, Yang, J, Liu, J.F. | | Deposit date: | 2017-12-28 | | Release date: | 2018-09-05 | | Last modified: | 2018-10-03 | | Method: | X-RAY DIFFRACTION (1.661 Å) | | Cite: | A positive-charged patch and stabilized hydrophobic core are essential for avirulence function of AvrPib in the rice blast fungus.
Plant J., 96, 2018
|
|
5ZK7
 
 | | Stapled-peptides tailored against initiation of translation | | Descriptor: | 7N-METHYL-8-HYDROGUANOSINE-5'-TRIPHOSPHATE, ACE-ARG-TYR-SER-ARG-MK8-GLN-LEU-LEU-MK8-LEU-PHE-ARG-NH2, CHLORIDE ION, ... | | Authors: | Lama, D, Liberator, A, Frosi, Y, Nakhle, J, Tsomia, N, Bashir, T, Lane, D.P, Brown, C.J, Verma, C.S, Auvin, S, Ciesielski, F, Uhring, M. | | Deposit date: | 2018-03-23 | | Release date: | 2019-02-20 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.12 Å) | | Cite: | Structural insights reveal a recognition feature for tailoring hydrocarbon stapled-peptides against the eukaryotic translation initiation factor 4E protein.
Chem Sci, 10, 2019
|
|
5ZML
 
 | | Stapled-peptides tailored against initiation of translation | | Descriptor: | 1,2-ETHANEDIOL, ACE-LYS-LYS-ARG-TYR-SER-ARG-MK8-GLN-LEU-LEU-MK8-PHE-ARG-ARG, Eukaryotic translation initiation factor 4E, ... | | Authors: | Lama, D, Liberator, A, Frosi, Y, Nakhle, J, Tsomia, N, Bashir, T, Lane, D.P, Brown, C.J, Verma, C.S, Auvin, S, Yano, J. | | Deposit date: | 2018-04-04 | | Release date: | 2019-02-20 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural insights reveal a recognition feature for tailoring hydrocarbon stapled-peptides against the eukaryotic translation initiation factor 4E protein.
Chem Sci, 10, 2019
|
|
2GYI
 
 | | DESIGN, SYNTHESIS, AND CHARACTERIZATION OF A POTENT XYLOSE ISOMERASE INHIBITOR, D-THREONOHYDROXAMIC ACID, AND HIGH-RESOLUTION X-RAY CRYSTALLOGRAPHIC STRUCTURE OF THE ENZYME-INHIBITOR COMPLEX | | Descriptor: | 2,3,4,N-TETRAHYDROXY-BUTYRIMIDIC ACID, MAGNESIUM ION, XYLOSE ISOMERASE | | Authors: | Allen, K.N, Lavie, A, Petsko, G.A, Ringe, D. | | Deposit date: | 1994-09-01 | | Release date: | 1995-07-10 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Design, Synthesis, and Characterization of a Potent Xylose Isomerase Inhibitor, D-Threonohydroxamic Acid, and High-Resolution X-Ray Crystallographic Structure of the Enzyme-Inhibitor Complex
Biochemistry, 34, 1995
|
|
5ZJY
 
 | | Stapled-peptides tailored against initiation of translation | | Descriptor: | 7-METHYL-GUANOSINE-5'-TRIPHOSPHATE, Eukaryotic translation initiation factor 4E, LYS-LYS-ARG-TYR-SER-ARG-2JN-GLN-LEU-LEU-2JN-PHE | | Authors: | Lama, D, Liberator, A, Frosi, Y, Nakhle, J, Tsomia, N, Bashir, T, Lane, D.P, Brown, C.J, Verma, C.S, Auvin, S, Ciesielski, B, Uhring, M. | | Deposit date: | 2018-03-22 | | Release date: | 2019-03-20 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.59 Å) | | Cite: | Structural insights reveal a recognition feature for tailoring hydrocarbon stapled-peptides against the eukaryotic translation initiation factor 4E protein.
Chem Sci, 10, 2019
|
|
5ZK5
 
 | | Stapled-peptides tailored against initiation of translation | | Descriptor: | 7N-METHYL-8-HYDROGUANOSINE-5'-TRIPHOSPHATE, Eukaryotic translation initiation factor 4E, LYS-ARG-TYR-SER-ARG-GLU-GLN-LEU-LEU-MK8-PHE-GLN-ARG-MK8 | | Authors: | Lama, D, Liberator, A, Frosi, Y, Nakhle, J, Tsomia, N, Bashir, T, Lane, D.P, Brown, C.J, Verma, C.S, Auvin, S, Ciesielski, F, Uhring, M. | | Deposit date: | 2018-03-23 | | Release date: | 2019-02-20 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Structural insights reveal a recognition feature for tailoring hydrocarbon stapled-peptides against the eukaryotic translation initiation factor 4E protein.
Chem Sci, 10, 2019
|
|
4E61
 
 | |
5Z2I
 
 | | Structure of Dictyostelium discoideum mitochondrial calcium uniporter N-ternimal domain (Se-DdMCU-NTD) | | Descriptor: | Dictyostelium discoideum mitochondrial calcium uniporter | | Authors: | Yuan, Y, Wen, M, Chou, J, Li, D, Bo, O. | | Deposit date: | 2018-01-02 | | Release date: | 2019-01-02 | | Last modified: | 2020-07-15 | | Method: | X-RAY DIFFRACTION (2.141 Å) | | Cite: | Structural Characterization of the N-Terminal Domain of theDictyostelium discoideumMitochondrial Calcium Uniporter.
Acs Omega, 5, 2020
|
|
5ZGL
 
 | | hnRNP A1 segment GGGYGGS (residues 234-240) | | Descriptor: | 7-mer peptide from Heterogeneous nuclear ribonucleoprotein A1 | | Authors: | Xie, M, Luo, F, Gui, X, Zhao, M, He, J, Li, D, Liu, C. | | Deposit date: | 2018-03-09 | | Release date: | 2019-04-03 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (0.95 Å) | | Cite: | Structural basis for reversible amyloids of hnRNPA1 elucidates their role in stress granule assembly.
Nat Commun, 10, 2019
|
|
