8QIT
 
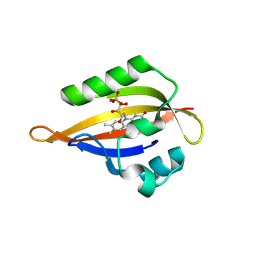 | | CrPhotLOV1 light state structure 77.5 ms (75-80 ms) after illumination determined by time-resolved serial synchrotron crystallography at room temperature | | Descriptor: | FLAVIN MONONUCLEOTIDE, Phototropin | | Authors: | Gotthard, G, Mous, S, Weinert, T, Maia, R.N.A, James, D, Dworkowski, F, Gashi, D, Antonia, F, Wang, M, Panepucci, E, Ozerov, D, Schertler, G.F.X, Heberle, J, Standfuss, J, Nogly, P. | | Deposit date: | 2023-09-12 | | Release date: | 2024-07-24 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Capturing the blue-light activated state of the Phot-LOV1 domain from Chlamydomonas reinhardtii using time-resolved serial synchrotron crystallography.
Iucrj, 11, 2024
|
|
6MBU
 
 | | Crystal structure of wild-type KRAS (1-169) bound to GDP and Mg (Space group P3) | | Descriptor: | GLYCEROL, GTPase KRas, GUANOSINE-5'-DIPHOSPHATE, ... | | Authors: | Dharmaiah, S, Tran, T.H, Yan, W, Simanshu, D.K. | | Deposit date: | 2018-08-30 | | Release date: | 2019-07-31 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Structures of N-terminally processed KRAS provide insight into the role of N-acetylation.
Sci Rep, 9, 2019
|
|
6TTG
 
 | | Crystal structure of the ATP binding domain of S. aureus GyrB complexed with LMD62 | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 2-[[3,4-bis(chloranyl)-5-methyl-1~{H}-pyrrol-2-yl]carbonylamino]-4-(2-morpholin-4-ylethoxy)-1,3-benzothiazole-6-carboxylic acid, CALCIUM ION, ... | | Authors: | Welin, M, Kimbung, R, Focht, D. | | Deposit date: | 2019-12-27 | | Release date: | 2020-12-30 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | New dual ATP-competitive inhibitors of bacterial DNA gyrase and topoisomerase IV active against ESKAPE pathogens.
Eur.J.Med.Chem., 213, 2021
|
|
6TOT
 
 | | Crystal structure of the Orexin-1 receptor in complex with lemborexant | | Descriptor: | (1R)-2-{[(S)-{[(2S)-2,3-dihydroxypropyl]oxy}(hydroxy)phosphoryl]oxy}-1-[(hexadecanoyloxy)methyl]ethyl (9Z)-octadec-9-enoate, (1~{R},2~{S})-2-[(2,4-dimethylpyrimidin-5-yl)oxymethyl]-~{N}-(5-fluoranylpyridin-2-yl)-2-(3-fluorophenyl)cyclopropane-1-carboxamide, Orexin receptor type 1, ... | | Authors: | Rappas, M, Ali, A, Bennett, K.A, Brown, J.D, Bucknell, S.J, Congreve, M, Cooke, R.M, Cseke, G, de Graaf, C, Dore, A.S, Errey, J.C, Jazayeri, A, Marshall, F.H, Mason, J.S, Mould, R, Patel, J.C, Tehan, B.G, Weir, M, Christopher, J.A. | | Deposit date: | 2019-12-11 | | Release date: | 2020-01-15 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.22 Å) | | Cite: | Comparison of Orexin 1 and Orexin 2 Ligand Binding Modes Using X-ray Crystallography and Computational Analysis.
J.Med.Chem., 63, 2020
|
|
7R3K
 
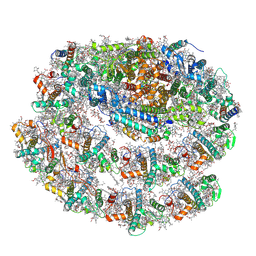 | | Chlamydomonas reinhardtii TSP9 mutant small Photosystem I complex | | Descriptor: | (1~{S})-3,5,5-trimethyl-4-[(1~{E},3~{E},5~{E},7~{E},9~{E},11~{E},13~{E},15~{E},17~{E})-3,7,12,16-tetramethyl-18-[(4~{S})-2,6,6-trimethyl-4-oxidanyl-cyclohexen-1-yl]octadeca-1,3,5,7,9,11,13,15,17-nonaenyl]cyclohex-3-en-1-ol, (3R)-beta,beta-caroten-3-ol, (3R,3'R,6S)-4,5-DIDEHYDRO-5,6-DIHYDRO-BETA,BETA-CAROTENE-3,3'-DIOL, ... | | Authors: | Klaiman, D, Schwartz, T, Nelson, N. | | Deposit date: | 2022-02-07 | | Release date: | 2023-02-22 | | Last modified: | 2023-04-12 | | Method: | ELECTRON MICROSCOPY (2.52 Å) | | Cite: | Structure of Photosystem I Supercomplex Isolated from a Chlamydomonas reinhardtii Cytochrome b6f Temperature-Sensitive Mutant.
Biomolecules, 13, 2023
|
|
5NVL
 
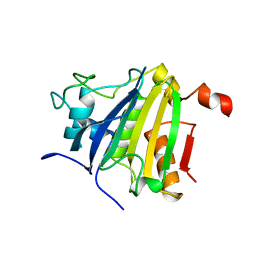 | | Crystal structure of the human 4EHP-GIGYF2 complex | | Descriptor: | Eukaryotic translation initiation factor 4E type 2, GRB10-interacting GYF protein 2 | | Authors: | Peter, D, Valkov, E. | | Deposit date: | 2017-05-04 | | Release date: | 2017-07-26 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | GIGYF1/2 proteins use auxiliary sequences to selectively bind to 4EHP and repress target mRNA expression.
Genes Dev., 31, 2017
|
|
6Z2I
 
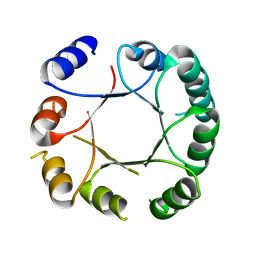 | | Crystal structure of DeNovoTIM6, a de novo designed TIM barrel | | Descriptor: | de novo designed TIM barrel DeNovoTIM6 | | Authors: | Romero-Romero, S, Kordes, S, Shanmugaratnam, S, Rodriguez-Romero, A, Fernandez-Velasco, D.A, Hocker, B. | | Deposit date: | 2020-05-15 | | Release date: | 2021-07-21 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.901 Å) | | Cite: | The Stability Landscape of de novo TIM Barrels Explored by a Modular Design Approach.
J.Mol.Biol., 433, 2021
|
|
1B2C
 
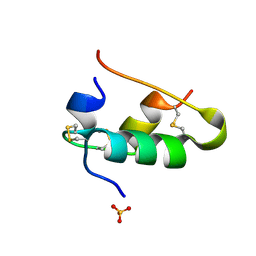 | |
6E4L
 
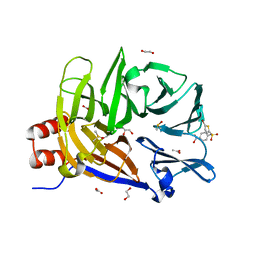 | | The structure of the N-terminal domain of human clathrin heavy chain 1 (nTD) in complex with ES9 | | Descriptor: | 1,2-ETHANEDIOL, 5-bromo-N-(4-nitrophenyl)thiophene-2-sulfonamide, ACETATE ION, ... | | Authors: | Dejonghe, W, Sharma, I, Denoo, B, Munck, S.D, Bulut, H, Mylle, E, Vasileva, M, Lu, Q, Savatin, D.V, Mishev, K, Nerinckx, W, Staes, A, Drozdzecki, A, Audenaert, D, Madder, A, Friml, J, Damme, D.V, Gevaert, K, Haucke, V, Savvides, S, Winne, J, Russinova, E. | | Deposit date: | 2018-07-17 | | Release date: | 2019-04-24 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Disruption of endocytosis through chemical inhibition of clathrin heavy chain function.
Nat.Chem.Biol., 15, 2019
|
|
6CWA
 
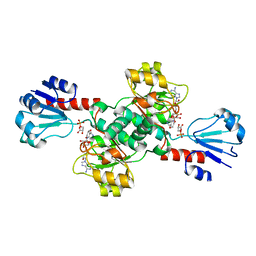 | |
6R60
 
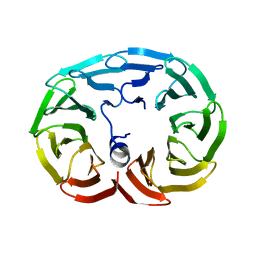 | |
6TP3
 
 | | Crystal structure of the Orexin-1 receptor in complex with daridorexant | | Descriptor: | (1R)-2-{[(S)-{[(2S)-2,3-dihydroxypropyl]oxy}(hydroxy)phosphoryl]oxy}-1-[(hexadecanoyloxy)methyl]ethyl (9Z)-octadec-9-enoate, Orexin receptor type 1, SULFATE ION, ... | | Authors: | Rappas, M, Ali, A, Bennett, K.A, Brown, J.D, Bucknell, S.J, Congreve, M, Cooke, R.M, Cseke, G, de Graaf, C, Dore, A.S, Errey, J.C, Jazayeri, A, Marshall, F.H, Mason, J.S, Mould, R, Patel, J.C, Tehan, B.G, Weir, M, Christopher, J.A. | | Deposit date: | 2019-12-12 | | Release date: | 2020-01-01 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (3.04 Å) | | Cite: | Comparison of Orexin 1 and Orexin 2 Ligand Binding Modes Using X-ray Crystallography and Computational Analysis.
J.Med.Chem., 63, 2020
|
|
3NP1
 
 | | CRYSTAL STRUCTURE OF THE COMPLEX OF NITROPHORIN 1 FROM RHODNIUS PROLIXUS WITH CYANIDE | | Descriptor: | CYANIDE ION, NITROPHORIN 1, PHOSPHATE ION, ... | | Authors: | Weichsel, A, Andersen, J.F, Champagne, D.E, Walker, F.A, Montfort, W.R. | | Deposit date: | 1998-01-22 | | Release date: | 1998-05-27 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structures of a nitric oxide transport protein from a blood-sucking insect.
Nat.Struct.Biol., 5, 1998
|
|
4UEL
 
 | | UCH-L5 in complex with ubiquitin-propargyl bound to the RPN13 DEUBAD domain | | Descriptor: | POLYUBIQUITIN-B, PROTEASOMAL UBIQUITIN RECEPTOR ADRM1, UBIQUITIN CARBOXYL-TERMINAL HYDROLASE ISOZYME L5 | | Authors: | Sahtoe, D.D, Van Dijk, W.J, El Oualid, F, Ekkebus, R, Ovaa, H, Sixma, T.K. | | Deposit date: | 2014-12-18 | | Release date: | 2015-03-04 | | Last modified: | 2019-04-03 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Mechanism of Uch-L5 Activation and Inhibition by Deubad Domains in Rpn13 and Ino80G.
Mol.Cell, 57, 2015
|
|
4XOU
 
 | | Crystal structure of the SR Ca2+-ATPase in the Ca2-E1-MgAMPPCP form determined by serial femtosecond crystallography using an X-ray free-electron laser. | | Descriptor: | CALCIUM ION, PHOSPHOMETHYLPHOSPHONIC ACID ADENYLATE ESTER, POTASSIUM ION, ... | | Authors: | Bublitz, M, Nass, K, Drachmann, N.D, Markvardsen, A.J, Gutmann, M.J, Barends, T.R.M, Mattle, D, Shoeman, R.L, Doak, R.B, Boutet, S, Messerschmidt, M, Seibert, M.M, Williams, G.J, Foucar, L, Reinhard, L, Sitsel, O, Gregersen, J.L, Clausen, J.D, Boesen, T, Gotfryd, K, Wang, K.-T, Olesen, C, Moller, J.V, Nissen, P, Schlichting, I. | | Deposit date: | 2015-01-16 | | Release date: | 2015-06-10 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structural studies of P-type ATPase-ligand complexes using an X-ray free-electron laser.
Iucrj, 2, 2015
|
|
4UCT
 
 | | Fragment bound to H.influenza NAD dependent DNA ligase | | Descriptor: | 1-(2,4-dimethylbenzyl)-6-oxo-1,6-dihydropyridine-3-carboxamide, 2-amino-6-methyl-5-(propan-2-yloxy)-3H-[1,2,4]triazolo[1,5-a]pyrimidin-8-ium, DNA LIGASE | | Authors: | Hale, M, Brassington, C, Carcanague, D, Embrey, K, Eyermann, C.J, Giacobbe, R.A, Gingipali, L, Gowravaram, M, Harang, J, Howard, T, Ioannidis, G, Jahic, H, Kutschke, A, Laganas, V.A, Loch, J, Miller, M.D, Murphy-Benenato, K.E, Oguto, H, Otterbein, L, Patel, S.J, Shapiro, A.B, Boriack-Sjodin, P.A. | | Deposit date: | 2014-12-04 | | Release date: | 2015-10-14 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | From Fragments to Leads: Novel Bacterial Nad+-Dependent DNA Ligase Inhibitors
Tetrahedron Lett., 56, 2015
|
|
1B3O
 
 | | TERNARY COMPLEX OF HUMAN TYPE-II INOSINE MONOPHOSPHATE DEHYDROGENASE WITH 6-CL-IMP AND SELENAZOLE ADENINE DINUCLEOTIDE | | Descriptor: | 6-CHLOROPURINE RIBOSIDE, 5'-MONOPHOSPHATE, PROTEIN (INOSINE MONOPHOSPHATE DEHYDROGENASE 2), ... | | Authors: | Colby, T.D, Vanderveen, K, Strickler, M.D, Goldstein, B.M. | | Deposit date: | 1998-12-14 | | Release date: | 1999-04-12 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Crystal structure of human type II inosine monophosphate dehydrogenase: implications for ligand binding and drug design.
Proc.Natl.Acad.Sci.USA, 96, 1999
|
|
3NPV
 
 | | Optimization of the in silico designed Kemp eliminase KE70 by computational design and directed evolution | | Descriptor: | deoxyribose phosphate aldolase | | Authors: | Khersonsky, O, Rothlisberge, D, Wollacott, A.M, Dym, O, Baker, D, Tawfik, D.S, Israel Structural Proteomics Center (ISPC) | | Deposit date: | 2010-06-29 | | Release date: | 2011-02-09 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.48 Å) | | Cite: | Optimization of the in-silico-designed kemp eliminase KE70 by computational design and directed evolution
J.Mol.Biol., 407, 2011
|
|
4XQD
 
 | | X-ray structure analysis of xylanase-WT at pH4.0 | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Endo-1,4-beta-xylanase 2, IODIDE ION | | Authors: | Wan, Q, Park, J.M, Riccardi, D.M, Hanson, L.B, Fisher, Z, Smith, J.C, Ostermann, A, Schrader, T, Graham, D.E, Coates, L, Langan, P, Kovalevsky, A.Y. | | Deposit date: | 2015-01-19 | | Release date: | 2015-09-23 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Direct determination of protonation states and visualization of hydrogen bonding in a glycoside hydrolase with neutron crystallography.
Proc.Natl.Acad.Sci.USA, 112, 2015
|
|
6TO7
 
 | | Crystal structure of the Orexin-1 receptor in complex with suvorexant at 2.29 A resolution | | Descriptor: | (1R)-2-{[(S)-{[(2S)-2,3-dihydroxypropyl]oxy}(hydroxy)phosphoryl]oxy}-1-[(hexadecanoyloxy)methyl]ethyl (9Z)-octadec-9-enoate, CITRIC ACID, Orexin receptor type 1, ... | | Authors: | Rappas, M, Ali, A, Bennett, K.A, Brown, J.D, Bucknell, S.J, Congreve, M, Cooke, R.M, Cseke, G, de Graaf, C, Dore, A.S, Errey, J.C, Jazayeri, A, Marshall, F.H, Mason, J.S, Mould, R, Patel, J.C, Tehan, B.G, Weir, M, Christopher, J.A. | | Deposit date: | 2019-12-11 | | Release date: | 2020-01-01 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.26 Å) | | Cite: | Comparison of Orexin 1 and Orexin 2 Ligand Binding Modes Using X-ray Crystallography and Computational Analysis.
J.Med.Chem., 63, 2020
|
|
8PPR
 
 | |
5G3X
 
 | | Structure of recombinant granulovirus polyhedrin | | Descriptor: | GRANULOVIRUS POLYHEDRIN | | Authors: | Bunker, R.D, Chiu, E, Metcalf, P. | | Deposit date: | 2016-05-02 | | Release date: | 2017-02-22 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.66 Å) | | Cite: | Atomic structure of granulin determined from native nanocrystalline granulovirus using an X-ray free-electron laser.
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
6R9G
 
 | |
5GEP
 
 | | SULFITE REDUCTASE HEMOPROTEIN CARBON MONOXIDE COMPLEX REDUCED WITH CRII EDTA | | Descriptor: | CARBON MONOXIDE, IRON/SULFUR CLUSTER, POTASSIUM ION, ... | | Authors: | Crane, B.R, Getzoff, E.D. | | Deposit date: | 1997-07-10 | | Release date: | 1998-01-14 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Probing the catalytic mechanism of sulfite reductase by X-ray crystallography: structures of the Escherichia coli hemoprotein in complex with substrates, inhibitors, intermediates, and products.
Biochemistry, 36, 1997
|
|
6NWA
 
 | | The structure of the photosystem I IsiA super-complex | | Descriptor: | 1,2-DIPALMITOYL-PHOSPHATIDYL-GLYCEROLE, 1,2-DISTEAROYL-MONOGALACTOSYL-DIGLYCERIDE, BETA-CAROTENE, ... | | Authors: | Toporik, H, Li, J, Williams, D, Chiu, P.L, Mazor, Y. | | Deposit date: | 2019-02-06 | | Release date: | 2019-05-29 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (3.48 Å) | | Cite: | The structure of the stress-induced photosystem I-IsiA antenna supercomplex.
Nat.Struct.Mol.Biol., 26, 2019
|
|
