5AQR
 
 | | Fragment-based screening of HSP70 sheds light on the functional role of ATP-binding site residues | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 6-methoxyquinazolin-4-amine, BAG FAMILY MOLECULAR CHAPERONE REGULATOR 1, ... | | Authors: | Jones, A.M, Westwood, I.M, Osborne, J.D, Matthews, T.P, Cheeseman, M.D, Rowlands, M.G, Jeganathan, F, Burke, R, Lee, D, Kadi, N, Liu, M, Richards, M, McAndrew, C, Yahya, N, Dobson, S.E, Jones, K, Workman, P, Collins, I, van Montfort, R.L.M. | | Deposit date: | 2015-09-22 | | Release date: | 2016-10-05 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.91 Å) | | Cite: | A fragment-based approach applied to a highly flexible target: Insights and challenges towards the inhibition of HSP70 isoforms.
Sci Rep, 6, 2016
|
|
6AVL
 
 | | Orthorhombic Trypsin (295 K) in the presence of 50% xylose | | Descriptor: | BENZAMIDINE, CALCIUM ION, Cationic trypsin, ... | | Authors: | Juers, D.H. | | Deposit date: | 2017-09-02 | | Release date: | 2017-09-20 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The impact of cryosolution thermal contraction on proteins and protein crystals: volumes, conformation and order.
Acta Crystallogr D Struct Biol, 74, 2018
|
|
5BXA
 
 | | Structure of PslG from Pseudomonas aeruginosa in complex with mannose | | Descriptor: | 1,2-ETHANEDIOL, CADMIUM ION, CHLORIDE ION, ... | | Authors: | Baker, P, Little, D.J, Howell, P.L. | | Deposit date: | 2015-06-08 | | Release date: | 2015-10-07 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Characterization of the Pseudomonas aeruginosa Glycoside Hydrolase PslG Reveals That Its Levels Are Critical for Psl Polysaccharide Biosynthesis and Biofilm Formation.
J.Biol.Chem., 290, 2015
|
|
5W8H
 
 | | Crystal Structure of Lactate Dehydrogenase A in complex with inhibitor compound 11 | | Descriptor: | 2-[3-(4-fluorophenyl)-5-(trifluoromethyl)-1H-pyrazol-1-yl]-1,3-thiazole-4-carboxylic acid, ACETATE ION, DIMETHYL SULFOXIDE, ... | | Authors: | Lukacs, C.M, Dranow, D.M. | | Deposit date: | 2017-06-21 | | Release date: | 2018-01-17 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Discovery and Optimization of Potent, Cell-Active Pyrazole-Based Inhibitors of Lactate Dehydrogenase (LDH).
J. Med. Chem., 60, 2017
|
|
3HSY
 
 | | High resolution structure of a dimeric GluR2 N-terminal domain (NTD) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Glutamate receptor 2, SULFATE ION, ... | | Authors: | Rossmann, M, Sukumaran, M, Penn, A.C, Veprintsev, D.B, Greger, I.H. | | Deposit date: | 2009-06-11 | | Release date: | 2010-06-16 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Subunit-selective N-terminal domain associations organize the formation of AMPA receptor heteromers
Embo J., 30, 2011
|
|
6DWJ
 
 | | SAMHD1 Bound to Vidarabine-TP in the Catalytic Pocket | | Descriptor: | 2'-DEOXYADENOSINE 5'-TRIPHOSPHATE, 9-{5-O-[(S)-hydroxy{[(R)-hydroxy(phosphonooxy)phosphoryl]oxy}phosphoryl]-beta-D-arabinofuranosyl}-9H-purin-6-amine, Deoxynucleoside triphosphate triphosphohydrolase SAMHD1, ... | | Authors: | Knecht, K.M, Buzovetsky, O, Schneider, C, Thomas, D, Srikanth, V, Kaderali, L, Tofoleanu, F, Reiss, K, Ferreiros, N, Geisslinger, G, Batista, V.S, Ji, X, Cinatl, J, Keppler, O.T, Xiong, Y. | | Deposit date: | 2018-06-26 | | Release date: | 2018-10-24 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | The structural basis for cancer drug interactions with the catalytic and allosteric sites of SAMHD1.
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
5H9O
 
 | | Complex of Murine endoplasmic reticulum alpha-glucosidase II with D-Glucose | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, ... | | Authors: | Caputo, A.T, Roversi, P, Alonzi, D.S, Kiappes, J.L, Zitzmann, N. | | Deposit date: | 2015-12-29 | | Release date: | 2016-07-27 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.37 Å) | | Cite: | Structures of mammalian ER alpha-glucosidase II capture the binding modes of broad-spectrum iminosugar antivirals.
Proc.Natl.Acad.Sci.USA, 113, 2016
|
|
7LLI
 
 | | Stimulatory immune receptor protein complex | | Descriptor: | 1-deoxy-1-({2,6-dioxo-5-[(E)-propylideneamino]-1,2,3,6-tetrahydropyrimidin-4-yl}amino)-D-ribitol, Beta-2-microglobulin, Major histocompatibility complex class I-related gene protein, ... | | Authors: | Rice, M.T, Littler, D.R, Rossjohn, J, Gully, B.S. | | Deposit date: | 2021-02-04 | | Release date: | 2021-11-17 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Recognition of the antigen-presenting molecule MR1 by a V delta 3 + gamma delta T cell receptor.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
5ECE
 
 | | Tankyrase 1 with Phthalazinone 1 | | Descriptor: | 1,2-ETHANEDIOL, 2-[4-[3-[(4-oxidanylidene-3~{H}-phthalazin-1-yl)methyl]phenyl]carbonylpiperazin-1-yl]pyridine-3-carbonitrile, ACETATE ION, ... | | Authors: | Kazmirski, S.L, Johannes, J, Read, J.A, Howard, T, Larsen, N.A, Ferguson, A.D. | | Deposit date: | 2015-10-20 | | Release date: | 2015-11-25 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Discovery of AZ0108, an orally bioavailable phthalazinone PARP inhibitor that blocks centrosome clustering.
Bioorg.Med.Chem.Lett., 25, 2015
|
|
5FOQ
 
 | | Acetylcholinesterase in complex with C7653 | | Descriptor: | 2,5,8,11,14,17-HEXAOXANONADECAN-19-OL, 2-(2,4-dichlorophenoxy)-N-[4-(1-piperidinylmethyl)phenyl]acetamide, 2-(2-METHOXYETHOXY)ETHANOL, ... | | Authors: | Berg, L, Mishra, B.K, Andersson, D.C, Ekstrom, F, Linusson, A. | | Deposit date: | 2015-11-25 | | Release date: | 2016-03-02 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | The Nature of Activated Non-Classical Hydrogen Bonds: A Case Study on Acetylcholinesterase-Ligand Complexes.
Chemistry, 22, 2016
|
|
6DY0
 
 | | Rabbit N-acylethanolamine-hydrolyzing acid amidase (NAAA) covalently bound to beta-lactam inhibitor ARN726, in presence of Triton X-100 | | Descriptor: | (2S)-3-amino-2-{[(4-cyclohexylbutoxy)carbonyl]amino}propanethioic S-acid, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-{2-[4-(1,1,3,3-TETRAMETHYLBUTYL)PHENOXY]ETHOXY}ETHANOL, ... | | Authors: | Gorelik, A, Gebai, A, Illes, K, Piomelli, D, Nagar, B. | | Deposit date: | 2018-07-01 | | Release date: | 2018-09-26 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (3.014 Å) | | Cite: | Molecular mechanism of activation of the immunoregulatory amidase NAAA.
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
6D5P
 
 | |
5FSG
 
 | |
5DJQ
 
 | | The structure of CBB3 cytochrome oxidase. | | Descriptor: | CALCIUM ION, COPPER (II) ION, Cbb3-type cytochrome c oxidase subunit CcoN1, ... | | Authors: | Buschmann, S, Warkentin, E, Xie, H, Kohlstaedt, M, Langer, J.D, Ermler, U, Michel, H. | | Deposit date: | 2015-09-02 | | Release date: | 2016-01-13 | | Last modified: | 2016-07-27 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | The structure of cbb3 cytochrome oxidase provides insights into proton pumping.
Science, 329, 2010
|
|
3GWX
 
 | | MOLECULAR RECOGNITION OF FATTY ACIDS BY PEROXISOME PROLIFERATOR-ACTIVATED RECEPTORS | | Descriptor: | 5,8,11,14,17-EICOSAPENTAENOIC ACID, PROTEIN (PEROXISOME PROLIFERATOR ACTIVATED RECEPTOR (PPAR-DELTA)) | | Authors: | Xu, H.E, Lambert, M.H, Montana, V.G, Parks, D.J, Blanchard, S.G, Brown, P.J, Sternbach, D.D, Lehmann, J.M, Wisely, G.B, Willson, T.M, Kliewer, S.A, Milburn, M.V. | | Deposit date: | 1999-04-26 | | Release date: | 2000-04-26 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Molecular recognition of fatty acids by peroxisome proliferator-activated receptors.
Mol.Cell, 3, 1999
|
|
7LNY
 
 | |
6DO1
 
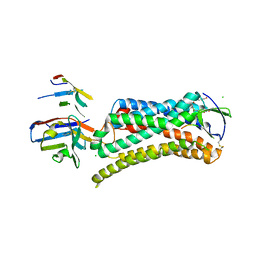 | | Structure of nanobody-stabilized angiotensin II type 1 receptor bound to S1I8 | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, 2-acetamido-2-deoxy-beta-D-glucopyranose, Angiotensin-like peptide S1I8, ... | | Authors: | Wingler, L.M, McMahon, C, Staus, D.P, Lefkowitz, R.J, Kruse, A.C. | | Deposit date: | 2018-06-08 | | Release date: | 2019-01-30 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.901 Å) | | Cite: | Distinctive Activation Mechanism for Angiotensin Receptor Revealed by a Synthetic Nanobody.
Cell, 176, 2019
|
|
6I2K
 
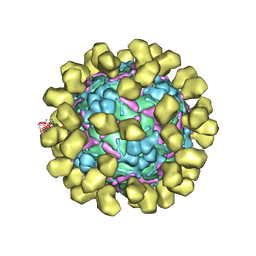 | | Structure of EV71 complexed with its receptor SCARB2 | | Descriptor: | 1-(2-aminopyridin-4-yl)-3-[(3S)-5-{4-[(E)-(ethoxyimino)methyl]phenoxy}-3-methylpentyl]imidazolidin-2-one, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Zhou, D, Zhao, Y, Kotecha, A, Fry, E.E, Kelly, J, Wang, X, Rao, Z, Rowlands, D.J, Ren, J, Stuart, D.I. | | Deposit date: | 2018-11-01 | | Release date: | 2018-11-28 | | Last modified: | 2020-07-29 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Unexpected mode of engagement between enterovirus 71 and its receptor SCARB2.
Nat Microbiol, 4, 2019
|
|
3K85
 
 | |
5T1J
 
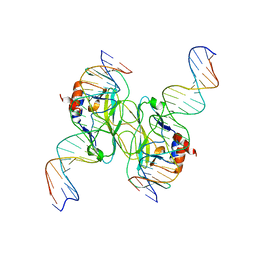 | | Crystal Structure of the Tbox DNA binding domain of the transcription factor T-bet | | Descriptor: | DNA, T-box transcription factor TBX21 | | Authors: | Liu, C.F, Brandt, G.S, Hoang, Q, Hwang, E.S, Naumova, N, Lazarevic, V, Dekker, J, Glimcher, L.H, Ringe, D, Petsko, G.A. | | Deposit date: | 2016-08-19 | | Release date: | 2016-10-26 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.947 Å) | | Cite: | Crystal structure of the DNA binding domain of the transcription factor T-bet suggests simultaneous recognition of distant genome sites.
Proc.Natl.Acad.Sci.USA, 113, 2016
|
|
3JWO
 
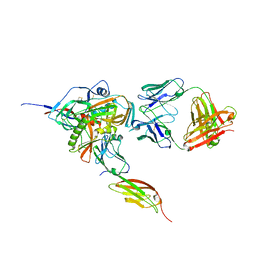 | | Structure of HIV-1 gp120 with gp41-Interactive Region: Layered Architecture and Basis of Conformational Mobility | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, FAB 48D Heavy CHAIN, FAB 48D LIGHT CHAIN, ... | | Authors: | Pancera, M, Majeed, S, Huang, C.C, Kwon, Y.D, Zhou, T, Robinson, J.E, Sodroski, J, Wyatt, R, Kwong, P.D. | | Deposit date: | 2009-09-18 | | Release date: | 2009-12-29 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (3.51 Å) | | Cite: | Structure of HIV-1 gp120 with gp41-interactive region reveals layered envelope architecture and basis of conformational mobility.
Proc.Natl.Acad.Sci.USA, 107, 2010
|
|
7XRP
 
 | | Cryo-EM structure of SARS-CoV-2 spike protein in complex with nanobody C5G2 (localized refinement) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, C5G2 nanobody, Spike protein S1 | | Authors: | Liu, L, Sun, H, Jiang, Y, Liu, X, Zhao, D, Zheng, Q, Li, S, Xia, N. | | Deposit date: | 2022-05-11 | | Release date: | 2022-10-05 | | Method: | ELECTRON MICROSCOPY (3.88 Å) | | Cite: | A potent synthetic nanobody with broad-spectrum activity neutralizes SARS-CoV-2 virus and the Omicron variant BA.1 through a unique binding mode.
J Nanobiotechnology, 20, 2022
|
|
6QG8
 
 | | Structure of human Bcl-2 in complex with PUMA BH3 peptide | | Descriptor: | Apoptosis regulator Bcl-2,Bcl-2-like protein 1,Apoptosis regulator Bcl-2,Bcl-2-like protein 1, Bcl-2-binding component 3 | | Authors: | Dokurno, P, Murray, J, Davidson, J, Chen, I, Davis, B, Graham, C.J, Harris, R, Jordan, A.M, Matassova, N, Pedder, C, Ray, S, Roughley, S, Smith, J, Walmsley, C, Wang, Y, Whitehead, N, Williamson, D.S, Casara, P, Le Diguarher, T, Hickman, J, Stark, J, Kotschy, A, Geneste, O, Hubbard, R.E. | | Deposit date: | 2019-01-10 | | Release date: | 2019-06-12 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Establishing Drug Discovery and Identification of Hit Series for the Anti-apoptotic Proteins, Bcl-2 and Mcl-1.
Acs Omega, 4, 2019
|
|
5ZUO
 
 | | Crystal Structure of BZ junction in diverse sequence | | Descriptor: | DNA (5'-D(*AP*CP*GP*GP*TP*TP*TP*AP*TP*CP*GP*CP*GP*CP*GP*CP*G)-3'), DNA (5'-D(*GP*TP*CP*GP*CP*GP*CP*GP*CP*GP*AP*TP*AP*AP*AP*CP*C)-3'), Double-stranded RNA-specific adenosine deaminase | | Authors: | Kim, K.K, Kim, D. | | Deposit date: | 2018-05-08 | | Release date: | 2018-08-29 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.902 Å) | | Cite: | Sequence preference and structural heterogeneity of BZ junctions.
Nucleic Acids Res., 46, 2018
|
|
8GCI
 
 | |
