7Q9E
 
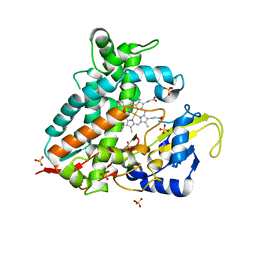 | | CYP106A1 | | Descriptor: | COBALT (II) ION, Cytochrome P450, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Carius, Y, Hutter, M, Kiss, F, Bernhardt, R, Lancaster, C.R.D. | | Deposit date: | 2021-11-12 | | Release date: | 2022-10-19 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structural comparison of the cytochrome P450 enzymes CYP106A1 and CYP106A2 provides insight into their differences in steroid conversion.
Febs Lett., 596, 2022
|
|
8UG1
 
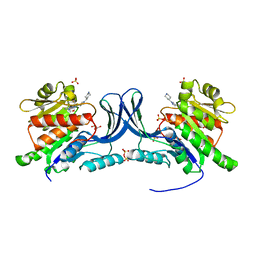 | | Crystal structure of KHK-C and compound 13 | | Descriptor: | GLYCEROL, Ketohexokinase, SULFATE ION, ... | | Authors: | Durbin, J.D, Guo, S.Y. | | Deposit date: | 2023-10-05 | | Release date: | 2023-12-06 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.99 Å) | | Cite: | Identification of LY3522348: A Highly Selective and Orally Efficacious Ketohexokinase Inhibitor.
J.Med.Chem., 66, 2023
|
|
6YRZ
 
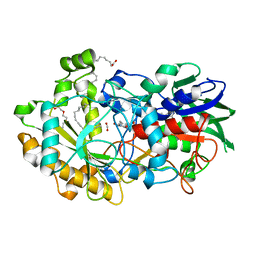 | | Crystal structure of FAP et pH 8.5 after illumination at 150K | | Descriptor: | CARBON DIOXIDE, FLAVIN-ADENINE DINUCLEOTIDE, Fatty acid photodecarboxylase, ... | | Authors: | Sorigue, D, Legrand, P, Blangy, S, Beisson, F, Arnoux, P. | | Deposit date: | 2020-04-20 | | Release date: | 2021-04-21 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.824 Å) | | Cite: | Mechanism and dynamics of fatty acid photodecarboxylase.
Science, 372, 2021
|
|
8TUC
 
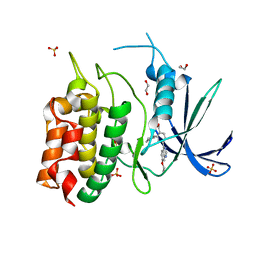 | | Unphosphorylated CaMKK2 in complex with CC-8977 | | Descriptor: | (4M)-2-cyclopentyl-4-(7-ethoxyquinazolin-4-yl)benzoic acid, 1,2-ETHANEDIOL, Calcium/calmodulin-dependent protein kinase kinase 2, ... | | Authors: | Bernard, S.M, Shanmugasundaram, V, D'Agostino, L. | | Deposit date: | 2023-08-16 | | Release date: | 2023-12-13 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Identification of Small Molecule Inhibitors and Ligand Directed Degraders of Calcium/Calmodulin Dependent Protein Kinase Kinase 1 and 2 (CaMKK1/2).
J.Med.Chem., 66, 2023
|
|
7O4X
 
 | |
5MTY
 
 | | Dibenzosuberone inhibitor 8e in complex with p38 MAPK | | Descriptor: | Mitogen-activated protein kinase 14, octyl beta-D-glucopyranoside, ~{N}-[2,4-bis(fluoranyl)-5-[[14-(2-hydroxyethylcarbamoyl)-2-oxidanylidene-6-tricyclo[9.4.0.0^{3,8}]pentadeca-1(15),3(8),4,6,11,13-hexaenyl]amino]phenyl]thiophene-2-carboxamide | | Authors: | Buehrmann, M, Rauh, D. | | Deposit date: | 2017-01-11 | | Release date: | 2017-09-06 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.31 Å) | | Cite: | Design, Synthesis, and Biological Evaluation of Novel Type I(1)/2 p38 alpha MAP Kinase Inhibitors with Excellent Selectivity, High Potency, and Prolonged Target Residence Time by Interfering with the R-Spine.
J. Med. Chem., 60, 2017
|
|
7T8J
 
 | | The ubiquitin-associated domain of human thirty-eight negative kinase-1 flexibly fused to the 1TEL crystallization chaperone via a GSGG linker | | Descriptor: | CHLORIDE ION, Transcription factor ETV6,Non-receptor tyrosine-protein kinase TNK1 | | Authors: | Soleimani, S, Pedroza Romo, M.J, Smith, T, Brown, S, Doukov, T, Moody, J.D. | | Deposit date: | 2021-12-16 | | Release date: | 2022-12-21 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.89 Å) | | Cite: | Fusion crystallization reveals the behavior of both the 1TEL crystallization chaperone and the TNK1 UBA domain.
Structure, 31, 2023
|
|
8FTM
 
 | | Setx-ssRNA-ADP-SO4 complex | | Descriptor: | 5'-3' RNA helicase-like protein, ADENOSINE-5'-DIPHOSPHATE, RNA (5'-R(*UP*UP*UP*UP*UP*UP*UP*UP*UP*UP*UP*U)-3'), ... | | Authors: | Williams, R.S, Appel, C.D. | | Deposit date: | 2023-01-12 | | Release date: | 2023-10-25 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (3.01 Å) | | Cite: | Sen1 architecture: RNA-DNA hybrid resolution, autoregulation, and insights into SETX inactivation in AOA2.
Mol.Cell, 83, 2023
|
|
6VCB
 
 | | Cryo-EM structure of the Glucagon-like peptide-1 receptor in complex with G protein, GLP-1 peptide and a positive allosteric modulator | | Descriptor: | 1-[(1R)-1-(2,6-dichloro-3-methoxyphenyl)ethyl]-6-{2-[(2R)-piperidin-2-yl]phenyl}-1H-benzimidazole, Glucagon-like peptide 1, Glucagon-like peptide 1 receptor, ... | | Authors: | Sun, B, Feng, D, Bueno, A, Kobilka, B, Sloop, K. | | Deposit date: | 2019-12-20 | | Release date: | 2020-07-22 | | Last modified: | 2020-09-30 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Structural insights into probe-dependent positive allosterism of the GLP-1 receptor.
Nat.Chem.Biol., 16, 2020
|
|
7OA7
 
 | | PilC minor pilin of Streptococcus sanguinis 2908 type IV pili | | Descriptor: | CALCIUM ION, PilC minor pilin, SODIUM ION | | Authors: | Sheppard, D, Pelicic, V. | | Deposit date: | 2021-04-19 | | Release date: | 2022-05-04 | | Last modified: | 2023-01-25 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Characterization of a glycan-binding complex of minor pilins completes the analysis of Streptococcus sanguinis type 4 pili subunits.
Proc.Natl.Acad.Sci.USA, 120, 2023
|
|
8PJF
 
 | | Human Leukocyte Antigen class II allotype DR1 presenting P11T->R modified influenza A virus haemagglutinin (HA)306-318 PKYVKQNTLKLAR | | Descriptor: | 1,2-ETHANEDIOL, HLA class II histocompatibility antigen, DR alpha chain, ... | | Authors: | MacLachlan, B.J, Wall, A, Greenshields-Watson, A.L, Cole, D.K, Rizkallah, P.J, Godkin, A.J. | | Deposit date: | 2023-06-23 | | Release date: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (1.48 Å) | | Cite: | A targeted single mutation in influenza A virus universal epitope transforms immunogenicity and protective immunity via CD4 + T cell activation.
Cell Rep, 43, 2024
|
|
6N9G
 
 | | Crystal Structure of RGS7-Gbeta5 dimer | | Descriptor: | Guanine nucleotide-binding protein subunit beta-5, Regulator of G-protein signaling 7 | | Authors: | Patil, D.N, Rangarajan, E, Izard, T, Martemyanov, K.A. | | Deposit date: | 2018-12-03 | | Release date: | 2019-01-09 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.129 Å) | | Cite: | Structural organization of a major neuronal G protein regulator, the RGS7-G beta 5-R7BP complex.
Elife, 7, 2018
|
|
6VGC
 
 | | Crystal Structures of FLAP bound to DG-031 | | Descriptor: | (2R)-cyclopentyl{4-[(quinolin-2-yl)methoxy]phenyl}acetic acid, 5-lipoxygenase-activating protein, CALCIUM ION, ... | | Authors: | Ho, J.D, Lee, M.R, Rauch, C.T, Aznavour, K, Park, J.S, Luz, J.G, Antonysamy, S, Condon, B, Maletic, M, Zhang, A, Hickey, M.J, Hughes, N.E, Chandrasekhar, S, Sloan, A.V, Gooding, K, Harvey, A, Yu, X.P, Kahl, S.D, Norman, B.H. | | Deposit date: | 2020-01-07 | | Release date: | 2020-12-02 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.37 Å) | | Cite: | Structure-based, multi-targeted drug discovery approach to eicosanoid inhibition: Dual inhibitors of mPGES-1 and 5-lipoxygenase activating protein (FLAP).
Biochim Biophys Acta Gen Subj, 1865, 2020
|
|
6PJJ
 
 | | Human PRPF4B bound to benzothiophene inhibitor 224 | | Descriptor: | 1,2-ETHANEDIOL, 4-(5-{[(3-aminophenyl)methyl]carbamoyl}thiophen-2-yl)-1-benzothiophene-2-carboxamide, PHOSPHATE ION, ... | | Authors: | Godoi, P.H.C, Santiago, A.S, Fala, A.M, Ramos, P.Z, Sriranganadane, D, Mascarello, A, Segretti, N, Azevedo, H, Guimaraes, C.R.W, Arruda, P, Elkins, J.M, Counago, R.M, Structural Genomics Consortium (SGC) | | Deposit date: | 2019-06-28 | | Release date: | 2019-08-28 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | to be published
To Be Published
|
|
6VBA
 
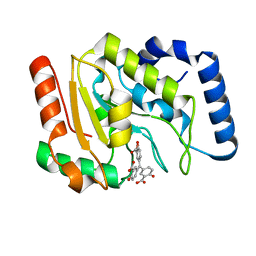 | | Structure of human Uracil DNA Glycosylase (UDG) bound to Aurintricarboxylic acid (ATA) | | Descriptor: | 3,3'-[(3-carboxy-4-oxocyclohexa-2,5-dien-1-ylidene)methylene]bis(6-hydroxybenzoic acid), Uracil-DNA glycosylase | | Authors: | Moiani, D, Arvai, A.S, Tainer, J.A. | | Deposit date: | 2019-12-18 | | Release date: | 2021-03-03 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | An effective human uracil-DNA glycosylase inhibitor targets the open pre-catalytic active site conformation.
Prog.Biophys.Mol.Biol., 163, 2021
|
|
5MSD
 
 | |
8FAR
 
 | |
6YMC
 
 | | 26-mer stem-loop RNA | | Descriptor: | BARIUM ION, RNA (26-MER) | | Authors: | Janowski, R, Niessing, D. | | Deposit date: | 2020-04-08 | | Release date: | 2022-04-20 | | Last modified: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Multiple intrinsically disordered RNA-binding motifs cooperate as RNA-folding catalyst and mediate phase transition
To Be Published
|
|
6F39
 
 | | C1r homodimer CUB1-EGF-CUB2 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, Complement C1r subcomponent, ... | | Authors: | Almitairi, J.O.M, Venkatraman Girija, U, Furze, C.M, Simpson-Gray, X, Badakshi, F, Marshall, J.E, Mitchell, D.A, Moody, P.C.E, Wallis, R. | | Deposit date: | 2017-11-28 | | Release date: | 2018-01-24 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (5.801 Å) | | Cite: | Structure of the C1r-C1s interaction of the C1 complex of complement activation.
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
6PLA
 
 | | Adducts formed after 1 month in the reaction of dichlorido(1,3-dimethylbenzimidazol-2-ylidene)(eta6-p-cymene)osmium(II) with HEWL | | Descriptor: | Lysozyme, OSMIUM ION, SODIUM ION, ... | | Authors: | Sullivan, M.P, Hartinger, C.G, Goldstone, D.C. | | Deposit date: | 2019-06-30 | | Release date: | 2020-11-25 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.25 Å) | | Cite: | Probing the Paradigm of Promiscuity for N-Heterocyclic Carbene Complexes and their Protein Adduct Formation.
Angew.Chem.Int.Ed.Engl., 2021
|
|
8Q2G
 
 | |
6PQY
 
 | | Cryo-EM structure of HzTransib/TIR DNA transposon end complex (TEC) | | Descriptor: | DNA (5'-D(P*CP*AP*CP*GP*GP*TP*GP*GP*AP*TP*CP*GP*AP*AP*AP*A)-3'), DNA (5'-D(P*TP*TP*TP*TP*CP*GP*AP*TP*CP*CP*AP*CP*CP*GP*TP*G)-3'), Putative DNA-mediated transposase | | Authors: | Liu, C, Yang, Y, Schatz, D.G. | | Deposit date: | 2019-07-10 | | Release date: | 2019-10-09 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (4.2 Å) | | Cite: | Structures of a RAG-like transposase during cut-and-paste transposition.
Nature, 575, 2019
|
|
4YEB
 
 | | Structural characterization of a synaptic adhesion complex | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, Fibronectin leucine rich transmembrane protein 3, ... | | Authors: | Ranaivoson, F.M, Liu, Q, Martini, F, Bergami, F, von Daake, S, Li, S, Lee, D, Demeler, B, Hendrickson, W.A, Comoletti, D. | | Deposit date: | 2015-02-23 | | Release date: | 2015-08-19 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (3.19 Å) | | Cite: | Structural and Mechanistic Insights into the Latrophilin3-FLRT3 Complex that Mediates Glutamatergic Synapse Development.
Structure, 23, 2015
|
|
6P7Q
 
 | | Structure of E. coli MS115-1 NucC, 5'-pApA bound form | | Descriptor: | CHLORIDE ION, E. coli MS115-1 NucC, RNA (5'-R(P*AP*A)-3') | | Authors: | Ye, Q, Lau, R.K, Berg, K.R, Corbett, K.D. | | Deposit date: | 2019-06-06 | | Release date: | 2019-12-25 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.66 Å) | | Cite: | Structure and Mechanism of a Cyclic Trinucleotide-Activated Bacterial Endonuclease Mediating Bacteriophage Immunity.
Mol.Cell, 77, 2020
|
|
7OFP
 
 | | Apo Structure of Mu2 Adaptin Subunit (Ap50) Of AP2 Clathrin Adaptor | | Descriptor: | AP-2 complex subunit mu, CITRIC ACID, GLYCEROL | | Authors: | Zaccai, N.R, Kelly, B.T, Evans, P.R, Owen, D.J. | | Deposit date: | 2021-05-05 | | Release date: | 2022-06-01 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.92 Å) | | Cite: | FCHO controls AP2's initiating role in endocytosis through a PtdIns(4,5)P 2 -dependent switch.
Sci Adv, 8, 2022
|
|
