6ZFO
 
 | | Association of two complexes of largely structurally disordered Spike ectodomain with bound EY6A Fab | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, EY6A heavy chain, EY6A light chain, ... | | Authors: | Duyvesteyn, H.M.E, Zhou, D, Zhao, Y, Fry, E.E, Ren, J, Stuart, D.I. | | Deposit date: | 2020-06-17 | | Release date: | 2020-07-08 | | Last modified: | 2021-12-22 | | Method: | ELECTRON MICROSCOPY (4.4 Å) | | Cite: | Structural basis for the neutralization of SARS-CoV-2 by an antibody from a convalescent patient.
Nat.Struct.Mol.Biol., 27, 2020
|
|
7U9G
 
 | | Rabies virus glycoprotein pre-fusion trimer in complex with neutralizing antibody RVA122 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Glycoprotein, RVA122 Fab Heavy Chain, ... | | Authors: | Callaway, H.M, Zyla, D, Larrous, F, Dias de Melo, G, Hastie, K.M, Avalos, R.D, Agarwal, A, Bouhry, H, Corti, D, Saphire, E.O. | | Deposit date: | 2022-03-10 | | Release date: | 2022-07-20 | | Method: | ELECTRON MICROSCOPY (3.39 Å) | | Cite: | Structure of the rabies virus glycoprotein trimer bound to a prefusion-specific neutralizing antibody.
Sci Adv, 8, 2022
|
|
6Z3K
 
 | | Structure of protective antibody 38-1-10A Fab | | Descriptor: | 1,2-ETHANEDIOL, Heavy chain, Light Chain | | Authors: | Zhou, D, Fry, E.E, Ren, J, Stuart, D.I. | | Deposit date: | 2020-05-20 | | Release date: | 2020-09-02 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structural and functional analysis of protective antibodies targeting the threefold plateau of enterovirus 71.
Nat Commun, 11, 2020
|
|
7UAB
 
 | | Human pro-meprin alpha (zymogen state) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Bayly-Jones, C, Lupton, C.J, Fritz, C, Schlenzig, D, Whisstock, J.C. | | Deposit date: | 2022-03-12 | | Release date: | 2022-11-02 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Helical ultrastructure of the metalloprotease meprin alpha in complex with a small molecule inhibitor.
Nat Commun, 13, 2022
|
|
6ZHD
 
 | | H11-H4 bound to Spike | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Nanobody H11-H4, ... | | Authors: | Clare, D.K, Naismith, J.H, Weckener, M, Vogirala, V.K. | | Deposit date: | 2020-06-22 | | Release date: | 2020-07-29 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | H11-H4 bound to Spike
To Be Published
|
|
8J12
 
 | | Cryo-EM structure of the AsCas12f-sgRNA-target DNA ternary complex | | Descriptor: | DNA (38-MER), MAGNESIUM ION, RNA (247-MER), ... | | Authors: | Hino, T, Omura, N.S, Nakagawa, R, Togashi, T, Takeda, N.S, Hiramoto, T, Tasaka, S, Hirano, H, Tokuyama, T, Uosaki, H, Ishiguro, H, Yamano, H, Ozaki, Y, Motooka, D, Mori, H, Kirita, Y, Kise, Y, Itoh, Y, Matoba, S, Aburatani, H, Yachie, N, Siksnys, V, Ohmori, T, Hoshino, A, Nureki, O. | | Deposit date: | 2023-04-12 | | Release date: | 2023-09-27 | | Method: | ELECTRON MICROSCOPY (3.08 Å) | | Cite: | Minimal and most efficient genome editing Cas enzyme
To Be Published
|
|
8J1J
 
 | | Cryo-EM structure of the AsCas12f-YHAM-sgRNAS3-5v7-target DNA | | Descriptor: | DNA (38-MER), MAGNESIUM ION, RNA (118-MER), ... | | Authors: | Hino, T, Omura, N.S, Nakagawa, R, Togashi, T, Takeda, N.S, Hiramoto, T, Tasaka, S, Hirano, H, Tokuyama, T, Uosaki, H, Ishiguro, H, Yamano, H, Ozaki, Y, Motooka, D, Mori, H, Kirita, Y, Kise, Y, Itoh, Y, Matoba, S, Aburatani, H, Yachie, N, Siksnys, V, Ohmori, T, Hoshino, A, Nureki, O. | | Deposit date: | 2023-04-13 | | Release date: | 2023-09-27 | | Method: | ELECTRON MICROSCOPY (2.91 Å) | | Cite: | Minimal and most efficient genome editing Cas enzyme
To Be Published
|
|
6Z43
 
 | | Cryo-EM Structure of SARS-CoV-2 Spike : H11-D4 Nanobody Complex | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Nanobody, ... | | Authors: | Ruza, R.R, Duyvesteyn, H.M.E, Shah, P, Carrique, L, Ren, J, Malinauskas, T, Zhou, D, Stuart, D.I, Naismith, J.H. | | Deposit date: | 2020-05-22 | | Release date: | 2020-06-03 | | Last modified: | 2020-07-29 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Structural basis for a potent neutralising single-domain antibody that blocks SARS-CoV-2 binding to its receptor ACE2
To Be Published
|
|
5N6V
 
 | | Crystal structure of Neisseria polysaccharea amylosucrase mutant derived from Neutral genetic Drift-based engineering | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, Amylosucrase, ... | | Authors: | Daude, D, Verges, A, Tranier, S. | | Deposit date: | 2017-02-16 | | Release date: | 2018-03-07 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Neutral Genetic Drift-Based Engineering of a Sucrose-Utilizing Enzyme toward Glycodiversification.
Acs Catalysis, 2019
|
|
8UTF
 
 | |
5NH7
 
 | |
5NHD
 
 | |
5NJQ
 
 | | Mix-and-diffuse serial synchrotron crystallography: structure of N,N',N''-Triacetylchitotriose bound to Lysozyme with 1s time-delay, phased with 4ET8 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CHLORIDE ION, Lysozyme C, ... | | Authors: | Oberthuer, D, Meents, A, Beyerlein, K.R, Chapman, H.N, Lieseke, J. | | Deposit date: | 2017-03-29 | | Release date: | 2017-10-18 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Mix-and-diffuse serial synchrotron crystallography.
IUCrJ, 4, 2017
|
|
5NJP
 
 | | Mix-and-diffuse serial synchrotron crystallography: structure of N,N',N''-Triacetylchitotriose bound to Lysozyme with 1s time-delay, phased with 1HEW | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CHLORIDE ION, Lysozyme C, ... | | Authors: | Oberthuer, D, Meents, A, Beyerlein, K.R, Chapman, H.N, Lieseke, J. | | Deposit date: | 2017-03-29 | | Release date: | 2017-10-18 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Mix-and-diffuse serial synchrotron crystallography.
IUCrJ, 4, 2017
|
|
5NRE
 
 | | A Native Ternary Complex of Alpha-1,3-Galactosyltransferase (a3GalT) Supports a Conserved Reaction Mechanism for Retaining Glycosyltransferases - a3GalT in complex with lactose - a3GalT-LAT | | Descriptor: | N-acetyllactosaminide alpha-1,3-galactosyltransferase, beta-D-galactopyranose-(1-4)-beta-D-glucopyranose | | Authors: | Albesa-Jove, D, Marina, A, Sainz-Polo, M.A, Guerin, M.E. | | Deposit date: | 2017-04-22 | | Release date: | 2017-10-11 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.98 Å) | | Cite: | Structural Snapshots of alpha-1,3-Galactosyltransferase with Native Substrates: Insight into the Catalytic Mechanism of Retaining Glycosyltransferases.
Angew. Chem. Int. Ed. Engl., 56, 2017
|
|
8ZH5
 
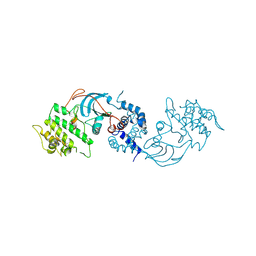 | |
7MUE
 
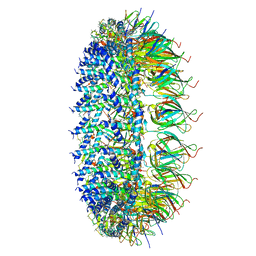 | | Legionella pneumophila Dot/Icm T4SS PR | | Descriptor: | DotF, IcmE protein, Type IV secretion protein IcmK, ... | | Authors: | Sheedlo, M.J, Durie, C.L, Swanson, M, Lacy, D.B, Ohi, M.D. | | Deposit date: | 2021-05-14 | | Release date: | 2021-10-06 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | Cryo-EM reveals new species-specific proteins and symmetry elements in the Legionella pneumophila Dot/Icm T4SS.
Elife, 10, 2021
|
|
8X7A
 
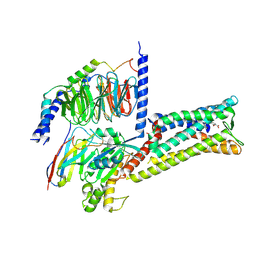 | | Treprostinil bound Prostacyclin Receptor G protein complex | | Descriptor: | 2-[[(1~{R},2~{R},3~{a}~{S},9~{a}~{S})-2-oxidanyl-1-[(3~{S})-3-oxidanyloctyl]-2,3,3~{a},4,9,9~{a}-hexahydro-1~{H}-cyclopenta[g]naphthalen-5-yl]oxy]ethanoic acid, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | Authors: | Wang, J.J, Jin, S, Zhang, H, Xu, Y, Hu, W, Jiang, Y, Chen, C, Wang, D.W, Xu, H.E, Wu, C. | | Deposit date: | 2023-11-23 | | Release date: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (2.56 Å) | | Cite: | Molecular recognition and activation of the prostacyclin receptor by anti-pulmonary arterial hypertension drugs.
Sci Adv, 10, 2024
|
|
8TKO
 
 | | KS-AT core of 6-deoxyerythronolide B synthase (DEBS) Module 3 crosslinked with its translocation ACP partner of Module 2 | | Descriptor: | EryAI, EryAII | | Authors: | Cogan, D.P, Soohoo, A.M, Chen, M, Brodsky, K.L, Liu, Y, Khosla, C. | | Deposit date: | 2023-07-25 | | Release date: | 2024-07-31 | | Method: | ELECTRON MICROSCOPY (3.05 Å) | | Cite: | Structural Basis for Intermodular Communication in Assembly-Line Polyketide Biosynthesis
To Be Published
|
|
9FWC
 
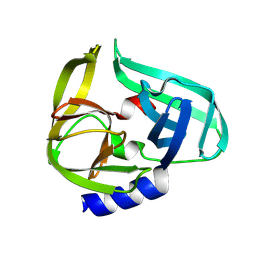 | | Coxsackievirus B3 3C protease in C121 spacegroup | | Descriptor: | Genome polyprotein | | Authors: | Fairhead, M, Lithgo, R.M, MacLean, E.M, Bowesman-Jones, H, Aschenbrenner, J.C, Balcomb, B.H, Capkin, E, Chandran, A.V, Godoy, A.S, Marples, P.G, Fearon, D, von Delft, F, Koekemoer, L. | | Deposit date: | 2024-06-28 | | Release date: | 2024-07-10 | | Method: | X-RAY DIFFRACTION (1.34 Å) | | Cite: | Coxsackievirus B3 3C protease in C121 spacegroup
To Be Published
|
|
8BNY
 
 | | Structure of the tetramerization domain of pLS20 conjugation repressor Rco | | Descriptor: | CHLORIDE ION, Immunity repressor protein | | Authors: | Bernardo, N, Crespo, I, Meijer, W.J.J, Boer, D.R. | | Deposit date: | 2022-11-14 | | Release date: | 2023-04-19 | | Last modified: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (1.429 Å) | | Cite: | A tetramerization domain in prokaryotic and eukaryotic transcription regulators homologous to p53.
Acta Crystallogr D Struct Biol, 79, 2023
|
|
6OB3
 
 | | Crystal structure of G13D-KRAS (GMPPNP-bound) in complex with GAP-related domain (GRD) of neurofibromin (NF1) | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, CHLORIDE ION, GTPase KRas, ... | | Authors: | Tran, T.H, Dharmaiah, S, Simanshu, D.K. | | Deposit date: | 2019-03-19 | | Release date: | 2019-10-16 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | KRAS G13D sensitivity to neurofibromin-mediated GTP hydrolysis.
Proc.Natl.Acad.Sci.USA, 116, 2019
|
|
9FS7
 
 | | Coxsackievirus A16 3C protease in C2 2 21 spacegroup | | Descriptor: | Genome polyprotein | | Authors: | Fairhead, M, Lithgo, R, MacLean, M, Bowesman-Jones, H, Aschenbrenner, J.C, Balcomb, B.H, Capkin, E, Chandran, A.V, Godoy, A.S, Marples, P.G, Fearon, D, von Delft, F, Koekemoer, L. | | Deposit date: | 2024-06-20 | | Release date: | 2024-07-10 | | Method: | X-RAY DIFFRACTION (1.37 Å) | | Cite: | Coxsackievirus A16 3C protease in C2 2 21 spacegroup
To Be Published
|
|
9PCY
 
 | | HIGH-RESOLUTION SOLUTION STRUCTURE OF REDUCED FRENCH BEAN PLASTOCYANIN AND COMPARISON WITH THE CRYSTAL STRUCTURE OF POPLAR PLASTOCYANIN | | Descriptor: | COPPER (II) ION, PLASTOCYANIN | | Authors: | Moore, J.M, Lepre, C.A, Gippert, G.P, Chazin, W.J, Case, D.A, Wright, P.E. | | Deposit date: | 1991-03-18 | | Release date: | 1993-10-31 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | High-resolution solution structure of reduced French bean plastocyanin and comparison with the crystal structure of poplar plastocyanin.
J.Mol.Biol., 221, 1991
|
|
8X5E
 
 | | Cryo-EM structure of human XPR1 in open state | | Descriptor: | PHOSPHATE ION, Solute carrier family 53 member 1, [(2~{R})-1-[2-azanylethoxy(oxidanyl)phosphoryl]oxy-3-hexadecanoyloxy-propan-2-yl] (~{Z})-octadec-9-enoate | | Authors: | Jiang, D.H, Yan, R. | | Deposit date: | 2023-11-17 | | Release date: | 2024-07-03 | | Method: | ELECTRON MICROSCOPY (3.61 Å) | | Cite: | Structure of transport at 3.6 Angstroms resolution.
To Be Published
|
|
