7KZ6
 
 | | Crystal structure of KabA from Bacillus cereus UW85 with bound cofactor PMP | | 分子名称: | 1,2-ETHANEDIOL, 4'-DEOXY-4'-AMINOPYRIDOXAL-5'-PHOSPHATE, Aminotransferase class I/II-fold pyridoxal phosphate-dependent enzyme | | 著者 | Prasertanan, T, Palmer, D.R.J, Sanders, D.A.R. | | 登録日 | 2020-12-10 | | 公開日 | 2021-05-26 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (1.65 Å) | | 主引用文献 | Snapshots along the catalytic path of KabA, a PLP-dependent aminotransferase required for kanosamine biosynthesis in Bacillus cereus UW85.
J.Struct.Biol., 213, 2021
|
|
6CQZ
 
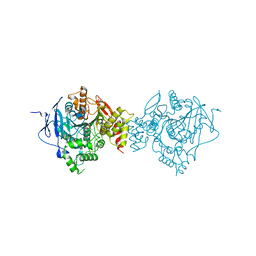 | | Crystal Structure of Recombinant Human Acetylcholinesterase Inhibited by VX | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, Acetylcholinesterase, ... | | 著者 | Bester, S.M, Guelta, M.A, Pegan, S.D, Height, J.J. | | 登録日 | 2018-03-16 | | 公開日 | 2018-12-05 | | 最終更新日 | 2023-10-04 | | 実験手法 | X-RAY DIFFRACTION (2.216 Å) | | 主引用文献 | Structural Insights of Stereospecific Inhibition of Human Acetylcholinesterase by VX and Subsequent Reactivation by HI-6.
Chem. Res. Toxicol., 31, 2018
|
|
7KZ5
 
 | | Crystal structure of KabA from Bacillus cereus UW85 in complex with the plp external aldimine adduct with kanosamine-6-phosphate | | 分子名称: | 1,2-ETHANEDIOL, 3-deoxy-3-[(E)-({3-hydroxy-2-methyl-5-[(phosphonooxy)methyl]pyridin-4-yl}methylidene)amino]-6-O-phosphono-alpha-D-gluco pyranose, Aminotransferase class I/II-fold pyridoxal phosphate-dependent enzyme, ... | | 著者 | Prasertanan, T, Palmer, D.R.J, Sanders, D.A.R. | | 登録日 | 2020-12-10 | | 公開日 | 2021-05-26 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (1.6 Å) | | 主引用文献 | Snapshots along the catalytic path of KabA, a PLP-dependent aminotransferase required for kanosamine biosynthesis in Bacillus cereus UW85.
J.Struct.Biol., 213, 2021
|
|
8DZA
 
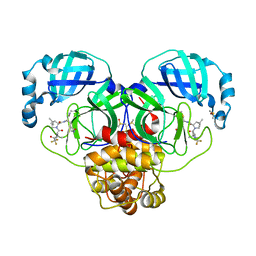 | | Crystal Structure of SARS-CoV-2 Main protease A193T mutant in complex with Nirmatrelvir | | 分子名称: | (1R,2S,5S)-N-{(1E,2S)-1-imino-3-[(3S)-2-oxopyrrolidin-3-yl]propan-2-yl}-6,6-dimethyl-3-[3-methyl-N-(trifluoroacetyl)-L-valyl]-3-azabicyclo[3.1.0]hexane-2-carboxamide, 3C-like proteinase nsp5, DIMETHYL SULFOXIDE | | 著者 | Noske, G.D, Oliva, G, Godoy, A.S. | | 登録日 | 2022-08-06 | | 公開日 | 2022-10-05 | | 最終更新日 | 2023-10-25 | | 実験手法 | X-RAY DIFFRACTION (1.961 Å) | | 主引用文献 | Structural basis of nirmatrelvir and ensitrelvir activity against naturally occurring polymorphisms of the SARS-CoV-2 main protease.
J.Biol.Chem., 299, 2023
|
|
7Z3U
 
 | | Crystal structure of SARS-CoV-2 Main Protease after incubation with Sulfo-Calpeptin | | 分子名称: | 3C-like proteinase nsp5, CHLORIDE ION, Calpetin, ... | | 著者 | Reinke, P.Y.A, Falke, S, Lieske, J, Ewert, W, Loboda, J, Rahmani Mashhour, A, Hauser, M, Karnicar, K, Usenik, A, Lindic, N, Lach, M, Boehler, H, Beck, T, Cox, R, Chapman, H.N, Hinrichs, W, Turk, D, Guenther, S, Meents, A. | | 登録日 | 2022-03-02 | | 公開日 | 2023-03-22 | | 最終更新日 | 2024-07-10 | | 実験手法 | X-RAY DIFFRACTION (1.72 Å) | | 主引用文献 | Calpeptin is a potent cathepsin inhibitor and drug candidate for SARS-CoV-2 infections.
Commun Biol, 6, 2023
|
|
7JWS
 
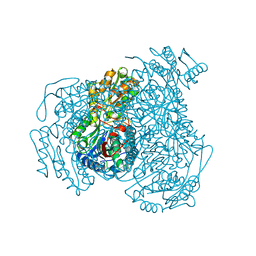 | | Crystal structure of human ALDH1A1 bound to compound (R)-28 | | 分子名称: | 1-methyl-5-phenyl-6-{[(1R)-1-phenylethyl]sulfanyl}-1,5-dihydro-4H-pyrazolo[3,4-d]pyrimidin-4-one, CHLORIDE ION, Retinal dehydrogenase 1, ... | | 著者 | Hurley, T.D, Buchman, C. | | 登録日 | 2020-08-26 | | 公開日 | 2020-12-30 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (1.6 Å) | | 主引用文献 | Development of 2,5-dihydro-4H-pyrazolo[3,4-d]pyrimidin-4-one inhibitors of aldehyde dehydrogenase 1A (ALDH1A) as potential adjuncts to ovarian cancer chemotherapy.
Eur.J.Med.Chem., 211, 2020
|
|
7OKQ
 
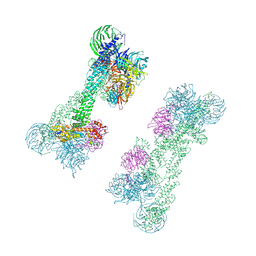 | | Cryo-EM Structure of the DDB1-DCAF1-CUL4A-RBX1 Complex | | 分子名称: | Cullin-4A, DDB1- and CUL4-associated factor 1, DNA damage-binding protein 1, ... | | 著者 | Mohamed, W.I, Schenk, A.D, Kempf, G, Cavadini, S, Thoma, N.H. | | 登録日 | 2021-05-18 | | 公開日 | 2021-10-13 | | 最終更新日 | 2024-07-10 | | 実験手法 | ELECTRON MICROSCOPY (8.4 Å) | | 主引用文献 | The CRL4 DCAF1 cullin-RING ubiquitin ligase is activated following a switch in oligomerization state.
Embo J., 40, 2021
|
|
8DZ6
 
 | | Crystal Structure of SARS-CoV-2 Main protease mutant Q189K in complex with Nirmatrelvir | | 分子名称: | (1R,2S,5S)-N-{(1E,2S)-1-imino-3-[(3S)-2-oxopyrrolidin-3-yl]propan-2-yl}-6,6-dimethyl-3-[3-methyl-N-(trifluoroacetyl)-L-valyl]-3-azabicyclo[3.1.0]hexane-2-carboxamide, 3C-like proteinase nsp5, DIMETHYL SULFOXIDE | | 著者 | Noske, G.D, Oliva, G, Godoy, A.S. | | 登録日 | 2022-08-06 | | 公開日 | 2022-10-05 | | 最終更新日 | 2023-10-25 | | 実験手法 | X-RAY DIFFRACTION (2.366 Å) | | 主引用文献 | Structural basis of nirmatrelvir and ensitrelvir activity against naturally occurring polymorphisms of the SARS-CoV-2 main protease.
J.Biol.Chem., 299, 2023
|
|
6O4F
 
 | | Structure of ALDH7A1 mutant N167S complexed with alpha-aminoadipate | | 分子名称: | 1,2-ETHANEDIOL, 2-AMINOHEXANEDIOIC ACID, Alpha-aminoadipic semialdehyde dehydrogenase | | 著者 | Tanner, J.J, Korasick, D.A, Laciak, A.R. | | 登録日 | 2019-02-28 | | 公開日 | 2019-07-24 | | 最終更新日 | 2023-11-15 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | Structural and biochemical consequences of pyridoxine-dependent epilepsy mutations that target the aldehyde binding site of aldehyde dehydrogenase ALDH7A1.
Febs J., 287, 2020
|
|
7ORW
 
 | | Non-structural protein 10 (nsp10) from SARS CoV-2 in complex with fragment VT00265 | | 分子名称: | 1H-benzimidazol-4-amine, CHLORIDE ION, DIMETHYL SULFOXIDE, ... | | 著者 | Talibov, V.O, Kozielski, F, Sele, C, Lou, J, Dong, D, Wang, Q, Shi, X, Nyblom, M, Rogstam, A, Krojer, T, Knecht, W, Fisher, S.Z. | | 登録日 | 2021-06-06 | | 公開日 | 2021-10-20 | | 最終更新日 | 2024-01-31 | | 実験手法 | X-RAY DIFFRACTION (1.95 Å) | | 主引用文献 | Identification of fragments binding to SARS-CoV-2 nsp10 reveals ligand-binding sites in conserved interfaces between nsp10 and nsp14/nsp16.
Rsc Chem Biol, 3, 2022
|
|
7TUD
 
 | | Crystal structure of HLA-B*44:05 (T73C) with 6mer EEFGRC and dipeptide GL | | 分子名称: | 1,2-ETHANEDIOL, Beta-2-microglobulin, EEFGRC peptide, ... | | 著者 | Jiang, J, Natarajan, K, Kim, E, Boyd, L.F, Margulies, D.H. | | 登録日 | 2022-02-02 | | 公開日 | 2022-09-07 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (1.45 Å) | | 主引用文献 | Structural mechanism of tapasin-mediated MHC-I peptide loading in antigen presentation.
Nat Commun, 13, 2022
|
|
8DZ2
 
 | | Crystal Structure of SARS-CoV-2 Main protease in complex with Nirmatrelvir | | 分子名称: | (1R,2S,5S)-N-{(1E,2S)-1-imino-3-[(3S)-2-oxopyrrolidin-3-yl]propan-2-yl}-6,6-dimethyl-3-[3-methyl-N-(trifluoroacetyl)-L-valyl]-3-azabicyclo[3.1.0]hexane-2-carboxamide, 3C-like proteinase nsp5, DIMETHYL SULFOXIDE | | 著者 | Noske, G.D, Oliva, G, Godoy, A.S. | | 登録日 | 2022-08-06 | | 公開日 | 2022-10-05 | | 最終更新日 | 2023-10-25 | | 実験手法 | X-RAY DIFFRACTION (2.129 Å) | | 主引用文献 | Structural basis of nirmatrelvir and ensitrelvir activity against naturally occurring polymorphisms of the SARS-CoV-2 main protease.
J.Biol.Chem., 299, 2023
|
|
7ORU
 
 | | Non-structural protein 10 (nsp10) from SARS CoV-2 in complex with fragment VT00221 | | 分子名称: | CHLORIDE ION, DIMETHYL SULFOXIDE, GLYCEROL, ... | | 著者 | Talibov, V.O, Kozielski, F, Sele, C, Lou, J, Dong, D, Wang, Q, Shi, X, Nyblom, M, Rogstam, A, Krojer, T, Knecht, W, Fisher, S.Z. | | 登録日 | 2021-06-06 | | 公開日 | 2021-10-20 | | 最終更新日 | 2024-01-31 | | 実験手法 | X-RAY DIFFRACTION (1.67 Å) | | 主引用文献 | Identification of fragments binding to SARS-CoV-2 nsp10 reveals ligand-binding sites in conserved interfaces between nsp10 and nsp14/nsp16.
Rsc Chem Biol, 3, 2022
|
|
6O6T
 
 | | Crystal structure of Csm6 H132A mutant | | 分子名称: | Csm6 | | 著者 | Jia, N, Patel, D.J. | | 登録日 | 2019-03-07 | | 公開日 | 2019-07-31 | | 最終更新日 | 2023-10-11 | | 実験手法 | X-RAY DIFFRACTION (2.31 Å) | | 主引用文献 | CRISPR-Cas III-A Csm6 CARF Domain Is a Ring Nuclease Triggering Stepwise cA4Cleavage with ApA>p Formation Terminating RNase Activity.
Mol.Cell, 75, 2019
|
|
6W7V
 
 | | Structure of rabbit actin in complex with truncated analog of Mycalolide B | | 分子名称: | (1E,3R,4R,5S,6R,9S,10S,12S)-12-[(4-aminobutanoyl)oxy]-1-[ethyl(formyl)amino]-4,10-dimethoxy-3,5,9,13-tetramethyltetradec-1-en-6-yl (2R)-oxolane-2-carboxylate, 1,2-ETHANEDIOL, ADENOSINE-5'-TRIPHOSPHATE, ... | | 著者 | Allingham, J.S, Deng, X, Trofimova, D. | | 登録日 | 2020-03-19 | | 公開日 | 2020-10-21 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (1.7 Å) | | 主引用文献 | Truncated Actin-Targeting Macrolide Derivative Blocks Cancer Cell Motility and Invasion of Extracellular Matrix.
J.Am.Chem.Soc., 143, 2021
|
|
6GUS
 
 | |
7TUC
 
 | | Crystal structure of HLA-B*44:05 (T73C) with 9mer EEFGRAFSF | | 分子名称: | 1,2-ETHANEDIOL, Beta-2-microglobulin, GLYCEROL, ... | | 著者 | Jiang, J, Natarajan, K, Kim, E, Boyd, L.F, Margulies, D.H. | | 登録日 | 2022-02-02 | | 公開日 | 2022-09-07 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (1.25 Å) | | 主引用文献 | Structural mechanism of tapasin-mediated MHC-I peptide loading in antigen presentation.
Nat Commun, 13, 2022
|
|
8E26
 
 | | Crystal Structure of SARS-CoV-2 Main Protease N142S mutant in complex with Nirmatrelvir | | 分子名称: | (1R,2S,5S)-N-{(1E,2S)-1-imino-3-[(3S)-2-oxopyrrolidin-3-yl]propan-2-yl}-6,6-dimethyl-3-[3-methyl-N-(trifluoroacetyl)-L-valyl]-3-azabicyclo[3.1.0]hexane-2-carboxamide, 3C-like proteinase nsp5, DIMETHYL SULFOXIDE | | 著者 | Noske, G.D, Godoy, A.S, Oliva, G. | | 登録日 | 2022-08-14 | | 公開日 | 2022-10-12 | | 最終更新日 | 2023-10-25 | | 実験手法 | X-RAY DIFFRACTION (1.845 Å) | | 主引用文献 | Structural basis of nirmatrelvir and ensitrelvir activity against naturally occurring polymorphisms of the SARS-CoV-2 main protease.
J.Biol.Chem., 299, 2023
|
|
8IKB
 
 | |
7TZ9
 
 | |
5V6X
 
 | |
5AFN
 
 | | alpha7-AChBP in complex with lobeline and fragment 5 | | 分子名称: | (4R)-4-(2-phenylethyl)pyrrolidin-2-one, 2-acetamido-2-deoxy-beta-D-glucopyranose, ACETYLCHOLINE-BINDING PROTEIN, ... | | 著者 | Spurny, R, Debaveye, S, Farinha, A, Veys, K, Gossas, T, Atack, J, Bertrand, D, Kemp, J, Vos, A, Danielson, U.H, Tresadern, G, Ulens, C. | | 登録日 | 2015-01-22 | | 公開日 | 2015-05-06 | | 最終更新日 | 2020-07-29 | | 実験手法 | X-RAY DIFFRACTION (2.149 Å) | | 主引用文献 | Molecular Blueprint of Allosteric Binding Sites in a Homologue of the Agonist-Binding Domain of the Alpha7 Nicotinic Acetylcholine Receptor.
Proc.Natl.Acad.Sci.USA, 112, 2015
|
|
7TZA
 
 | | Structure of PQS Response Protein PqsE in complex with N-(4-(3-neopentylureido)phenyl)-1H-indazole-7-carboxamide | | 分子名称: | 1,2-ETHANEDIOL, FE (III) ION, N-{4-[(2,2-dimethylpropyl)carbamamido]phenyl}-1H-indazole-7-carboxamide, ... | | 著者 | Jeffrey, P.D, Taylor, I.R, Bassler, B.L. | | 登録日 | 2022-02-15 | | 公開日 | 2022-08-31 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (2.1 Å) | | 主引用文献 | The PqsE Active Site as a Target for Small Molecule Antimicrobial Agents against Pseudomonas aeruginosa.
Biochemistry, 61, 2022
|
|
8OYR
 
 | | DNA Major Groove Binding by lambda-[Ru(phen)2(phi)]2+ | | 分子名称: | DNA (5'-D(*CP*CP*GP*GP*TP*AP*CP*CP*GP*G)-3'), STRONTIUM ION, ruthenium polypyridyl complex (lambda enantiomer) | | 著者 | Prieto-Otoya, T.D, Cardin, C.J, McQuaid, K.T, Paterson, N.G. | | 登録日 | 2023-05-05 | | 公開日 | 2024-03-13 | | 最終更新日 | 2024-03-27 | | 実験手法 | X-RAY DIFFRACTION (2.21 Å) | | 主引用文献 | Probing a Major DNA Weakness: Resolving the Groove and Sequence Selectivity of the Diimine Complex Lambda-[Ru(phen) 2 phi] 2.
Angew.Chem.Int.Ed.Engl., 63, 2024
|
|
6UGV
 
 | | Crystal structure of the Fab fragment of anti-TNFa antibody infliximab (Remicade) in a I-centered orthorhombic crystal form, Lot C | | 分子名称: | 1,2-ETHANEDIOL, Infliximab Fab Heavy Chain, Infliximab Fab Light Chain, ... | | 著者 | Lerch, T.F, Sharpe, P, Mayclin, S.J, Edwards, T.E, Polleck, S, Rouse, J.C, Conlan, H.D. | | 登録日 | 2019-09-26 | | 公開日 | 2019-11-13 | | 最終更新日 | 2023-10-11 | | 実験手法 | X-RAY DIFFRACTION (2.4 Å) | | 主引用文献 | Crystal Structures of PF-06438179/GP1111, an Infliximab Biosimilar.
BioDrugs, 34, 2020
|
|
