4LT7
 
 | | Crystal structure of the c2a domain of rabphilin-3a in complex with a calcium | | Descriptor: | CALCIUM ION, Rabphilin-3A | | Authors: | Verdaguer, N, Ferrer-Orta, C, Buxaderas, M, Corbalan-Garcia, S, Perez-Sanchez, D, Guerrero-Valero, M, Luengo, G, Pous, J, Guerra, P, Gomez-Fernandez, J.C, Guillen, J. | | Deposit date: | 2013-07-23 | | Release date: | 2013-12-11 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural insights into the Ca2+ and PI(4,5)P2 binding modes of the C2 domains of rabphilin 3A and synaptotagmin 1.
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
1U3Y
 
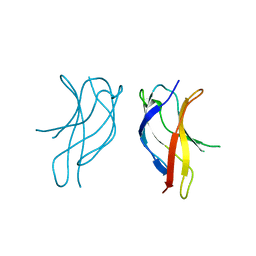 | | Crystal structure of ILAC mutant of dimerisation domain of NF-kB p50 transcription factor | | Descriptor: | Nuclear factor NF-kappa-B p105 subunit | | Authors: | Chirgadze, D.Y, Demydchuk, M, Becker, M, Moran, S, Paoli, M. | | Deposit date: | 2004-07-22 | | Release date: | 2004-08-17 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.901 Å) | | Cite: | Snapshot of Protein Structure Evolution Reveals Conservation of Functional Dimerization through Intertwined Folding
Structure, 12, 2004
|
|
1U42
 
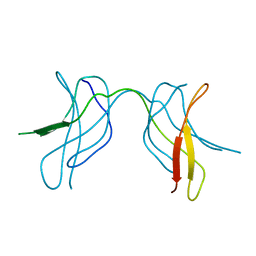 | | Crystal structure of MLAM mutant of dimerisation domain of NF-kB p50 transcription factor | | Descriptor: | Nuclear factor NF-kappa-B p105 subunit | | Authors: | Chirgadze, D.Y, Demydchuk, M, Becker, M, Moran, S, Paoli, M. | | Deposit date: | 2004-07-23 | | Release date: | 2004-08-17 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.699 Å) | | Cite: | Snapshot of Protein Structure Evolution Reveals Conservation of Functional Dimerization through Intertwined Folding
Structure, 12, 2004
|
|
1CRH
 
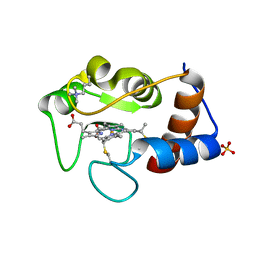 | |
8WJO
 
 | | Cryo-EM structure of 8-subunit Smc5/6 arm region | | Descriptor: | DNA repair protein KRE29, E3 SUMO-protein ligase MMS21, Structural maintenance of chromosomes protein 5, ... | | Authors: | Li, Q, Zhang, J, Zhang, X, Cheng, T, Wang, Z, Jin, D, Chen, Z, Wang, L. | | Deposit date: | 2023-09-26 | | Release date: | 2024-06-26 | | Last modified: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (6.04 Å) | | Cite: | Cryo-EM structures of Smc5/6 in multiple states reveal its assembly and functional mechanisms.
Nat.Struct.Mol.Biol., 31, 2024
|
|
1D1J
 
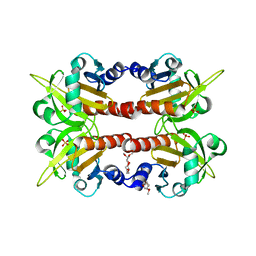 | | CRYSTAL STRUCTURE OF HUMAN PROFILIN II | | Descriptor: | 1-(2-METHOXY-ETHOXY)-2-{2-[2-(2-METHOXY-ETHOXY]-ETHOXY}-ETHANE, 1-METHOXY-2-[2-(2-METHOXY-ETHOXY]-ETHANE, PROFILIN II, ... | | Authors: | Nodelman, I.M, Bowman, G.D, Lindberg, U, Schutt, C.E. | | Deposit date: | 1999-09-17 | | Release date: | 2000-12-22 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | X-ray structure determination of human profilin II: A comparative structural analysis of human profilins.
J.Mol.Biol., 294, 1999
|
|
3EN3
 
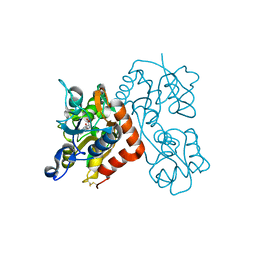 | | Crystal Structure of the GluR4 Ligand-Binding domain in complex with kainate | | Descriptor: | 3-(CARBOXYMETHYL)-4-ISOPROPENYLPROLINE, Glutamate receptor 4,Glutamate receptor | | Authors: | Gill, A, Madden, D.R. | | Deposit date: | 2008-09-25 | | Release date: | 2009-05-19 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.43 Å) | | Cite: | Correlating AMPA receptor activation and cleft closure across subunits: crystal structures of the GluR4 ligand-binding domain in complex with full and partial agonists
Biochemistry, 47, 2008
|
|
5ZUR
 
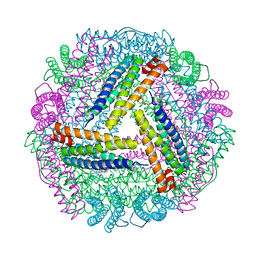 | | Achromobacter Dh1f Bacterioferritin | | Descriptor: | BARIUM ION, Bacterioferritin, CHLORIDE ION, ... | | Authors: | Dwivedy, A, Jha, B, Singh, K.H, Ahmed, M, Ashraf, A, Kumar, D, Biswal, B.K. | | Deposit date: | 2018-05-08 | | Release date: | 2018-09-05 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Serendipitous crystallization and structure determination of bacterioferritin from Achromobacter.
Acta Crystallogr F Struct Biol Commun, 74, 2018
|
|
8PJN
 
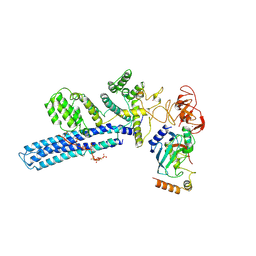 | | Catalytic module of human CTLH E3 ligase bound to multiphosphorylated UBE2H~ubiquitin | | Descriptor: | E3 ubiquitin-protein transferase MAEA, E3 ubiquitin-protein transferase RMND5A, Ubiquitin, ... | | Authors: | Chrustowicz, J, Sherpa, D, Prabu, R.J, Schulman, B.A. | | Deposit date: | 2023-06-23 | | Release date: | 2024-01-03 | | Last modified: | 2024-11-13 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Multisite phosphorylation dictates selective E2-E3 pairing as revealed by Ubc8/UBE2H-GID/CTLH assemblies.
Mol.Cell, 84, 2024
|
|
8PMQ
 
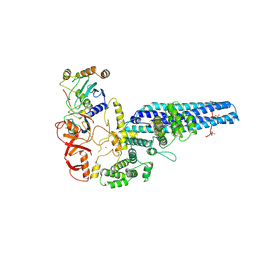 | | Catalytic module of yeast GID E3 ligase bound to multiphosphorylated Ubc8~ubiquitin | | Descriptor: | E3 ubiquitin-protein ligase RMD5, Protein FYV10, Ubiquitin, ... | | Authors: | Chrustowicz, J, Sherpa, D, Prabu, R.J, Schulman, B.A. | | Deposit date: | 2023-06-29 | | Release date: | 2024-01-03 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (3.53 Å) | | Cite: | Multisite phosphorylation dictates selective E2-E3 pairing as revealed by Ubc8/UBE2H-GID/CTLH assemblies.
Mol.Cell, 84, 2024
|
|
7TXL
 
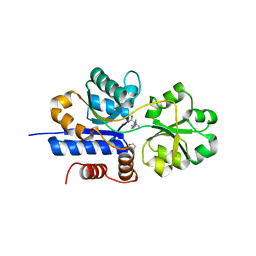 | | Crystal structure of EgtU solute binding domain from Streptococcus pneumoniae D39 in complex with L-ergothioneine | | Descriptor: | 1,2-ETHANEDIOL, Choline transporter (Glycine betaine transport system permease protein), trimethyl-[(2S)-1-oxidanyl-1-oxidanylidene-3-(2-sulfanylidene-1,3-dihydroimidazol-4-yl)propan-2-yl]azanium | | Authors: | Zhang, Y, Gonzalez-Gutierrez, G, Giedroc, D.P. | | Deposit date: | 2022-02-09 | | Release date: | 2022-12-21 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.44 Å) | | Cite: | Discovery and structure of a widespread bacterial ABC transporter specific for ergothioneine.
Nat Commun, 13, 2022
|
|
7TXK
 
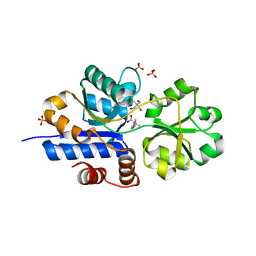 | | Crystal structure of EgtU solute binding domain from Streptococcus pneumoniae D39 in complex with L-ergothioneine | | Descriptor: | 1,2-ETHANEDIOL, Choline transporter (Glycine betaine transport system permease protein), SULFATE ION, ... | | Authors: | Zhang, Y, Gonzalez-Gutierrez, G, Giedroc, D.P. | | Deposit date: | 2022-02-09 | | Release date: | 2022-12-21 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.78 Å) | | Cite: | Discovery and structure of a widespread bacterial ABC transporter specific for ergothioneine.
Nat Commun, 13, 2022
|
|
1D7H
 
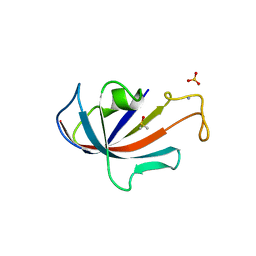 | | FKBP COMPLEXED WITH DMSO | | Descriptor: | AMMONIUM ION, DIMETHYL SULFOXIDE, PROTEIN (FK506-BINDING PROTEIN), ... | | Authors: | Burkhard, P, Taylor, P, Walkinshaw, M.D. | | Deposit date: | 1999-10-18 | | Release date: | 1999-10-21 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | X-ray structures of small ligand-FKBP complexes provide an estimate for hydrophobic interaction energies.
J.Mol.Biol., 295, 2000
|
|
3IXY
 
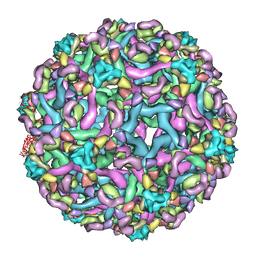 | | The pseudo-atomic structure of dengue immature virus in complex with Fab fragments of the anti-fusion loop antibody E53 | | Descriptor: | E53 Fab Fragment (chain H), E53 Fab Fragment (chain L), Envelope protein E, ... | | Authors: | Cherrier, M.V, Kaufmann, B, Nybakken, G.E, Lok, S.M, Warren, J.T, Nelson, C.A, Kostyuchenko, V.A, Holdaway, H.A, Chipman, P.R, Kuhn, R.J, Diamond, M.S, Rossmann, M.G, Fremont, D.H. | | Deposit date: | 2009-02-26 | | Release date: | 2009-10-27 | | Last modified: | 2024-02-21 | | Method: | ELECTRON MICROSCOPY (23 Å) | | Cite: | Structural basis for the preferential recognition of immature flaviviruses by a fusion-loop antibody
Embo J., 28, 2009
|
|
4D2S
 
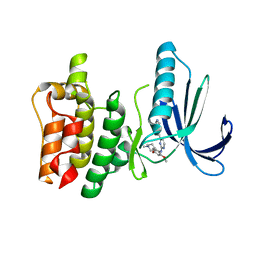 | | Human TTK in complex with a Dyrk1B inhibitor | | Descriptor: | DUAL SPECIFICITY PROTEIN KINASE TTK, N-{2-methoxy-4-[(1-methylpiperidin-4-yl)oxy]phenyl}-4-(1H-pyrrolo[2,3-c]pyridin-3-yl)pyrimidin-2-amine | | Authors: | Debreczeni, J.E, Kettle, J.G, Ballard, P, Bardelle, C, Butterworth, S, Colclough, N, Critchlow, S.E, Fairley, G, Fillery, S, Graham, M.A, Goodwin, L, Guichard, S, Hudson, K, Mahmood, A, Vincent, J, Ward, R.A, Whittaker, D. | | Deposit date: | 2014-05-12 | | Release date: | 2015-04-22 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Discovery and Optimization of a Novel Series of Dyrk1B Kinase Inhibitors to Explore a Mek Resistance Hypothesis.
J.Med.Chem., 58, 2015
|
|
8FH5
 
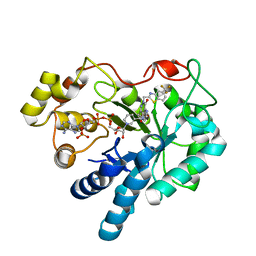 | | Crystal Structure Of Aldose Reductase (AKR1B1) Complexed With NADP+ And AT-001 | | Descriptor: | (8-oxo-7-{[5-(trifluoromethyl)-1,3-benzothiazol-2-yl]methyl}-7,8-dihydropyrazino[2,3-d]pyridazin-5-yl)acetic acid, Aldo-keto reductase family 1 member B1, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Arenas, R, Wilson, D.K. | | Deposit date: | 2022-12-13 | | Release date: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.62 Å) | | Cite: | Crystal Structure Of Aldose Reductase (AKR1B1) Complexed With NADP+ And AT-001
To Be Published
|
|
5F02
 
 | | CATHEPSIN L IN COMPLEX WITH (2S,4R)-4-(2-Chloro-4-methoxy-benzenesulfonyl)-1-[3-(5-chloro-pyridin-2-yl)-azetidine-3-carbonyl]-pyrrolidine-2-carboxylic acid (1-cyano-cyclopropyl)-amide | | Descriptor: | (2~{S},4~{R})-4-[(2-chloranyl-4-methoxy-phenyl)-bis(oxidanyl)-$l^{4}-sulfanyl]-1-[3-(5-chloranylpyridin-2-yl)azetidin-3-yl]carbonyl-~{N}-[1-(iminomethyl)cyclopropyl]pyrrolidine-2-carboxamide, Cathepsin L1, GLYCEROL | | Authors: | Banner, D, Benz, J, Stihle, M, Kuglstatter, A. | | Deposit date: | 2015-11-27 | | Release date: | 2016-02-24 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.43 Å) | | Cite: | A Real-World Perspective on Molecular Design.
J.Med.Chem., 59, 2016
|
|
8FH9
 
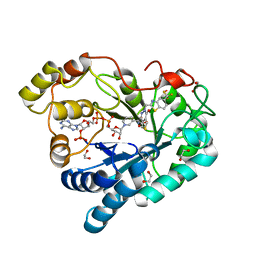 | | Crystal Structure Of Aldose Reductase (AKR1B1) Complexed With NADP+ And AT-007 | | Descriptor: | (4-oxo-3-{[5-(trifluoromethyl)-1,3-benzothiazol-2-yl]methyl}-3,4-dihydrothieno[3,4-d]pyridazin-1-yl)acetic acid, 1,2-ETHANEDIOL, Aldo-keto reductase family 1 member B1, ... | | Authors: | Arenas, R, Wilson, D.K. | | Deposit date: | 2022-12-13 | | Release date: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal Structure Of Aldose Reductase (AKR1B1) Complexed With NADP+ And AT-007
To Be Published
|
|
7M1M
 
 | | Crystal structure of Pseudomonas aeruginosa ClpP1 | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, ATP-dependent Clp protease proteolytic subunit | | Authors: | Mawla, G.D, Grant, R.A, Baker, T.A, Sauer, R.T. | | Deposit date: | 2021-03-13 | | Release date: | 2021-03-24 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | ClpP1P2 peptidase activity promotes biofilm formation in Pseudomonas aeruginosa.
Mol.Microbiol., 115, 2021
|
|
8A12
 
 | | Plasmodium falciparum Myosin A full-length, post-rigor state complexed to Mg.ATP-gamma-S | | Descriptor: | 1,2-ETHANEDIOL, ADENOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Moussaoui, D, Robblee, J.P, Auguin, D, Fisher, F, Fagnant, P.M, MacFarlane, J.E, Mueller-Dieckmann, C, Baum, J, Robert-Paganin, J, Trybus, K.M, Houdusse, A. | | Deposit date: | 2022-05-31 | | Release date: | 2023-06-21 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.03 Å) | | Cite: | Mechanism of small molecule inhibition of Plasmodium falciparum myosin A informs antimalarial drug design.
Nat Commun, 14, 2023
|
|
8FH6
 
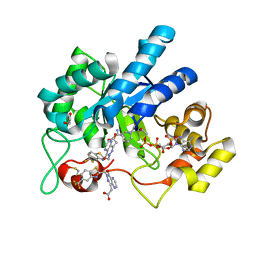 | | Crystal Structure Of Aldose Reductase (AKR1B1) Complexed With NADP+ And Two AT-001 | | Descriptor: | (8-oxo-7-{[5-(trifluoromethyl)-1,3-benzothiazol-2-yl]methyl}-7,8-dihydropyrazino[2,3-d]pyridazin-5-yl)acetic acid, 1,2-ETHANEDIOL, Aldo-keto reductase family 1 member B1, ... | | Authors: | Arenas, R, Wilson, D.K. | | Deposit date: | 2022-12-13 | | Release date: | 2023-12-20 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.952 Å) | | Cite: | Crystal Structure Of Aldose Reductase (AKR1B1) Complexed With NADP+ And Two AT-001
To Be Published
|
|
8FH7
 
 | |
6R8I
 
 | | PP4R3A EVH1 domain bound to FxxP motif | | Descriptor: | SER-LEU-PRO-PHE-THR-PHE-LYS-VAL-PRO-ALA-PRO-PRO-PRO-SER-LEU-PRO-PRO-SER, Serine/threonine-protein phosphatase 4 regulatory subunit 3A | | Authors: | Ueki, Y, Kruse, T, Weisser, M.B, Sundell, G.N, Yoo Larsen, M.S, Lopez Mendez, B, Jenkins, N.P, Garvanska, D.H, Cressey, L, Zhang, G, Davey, N, Montoya, G, Ivarsson, Y, Kettenbach, A, Nilsson, J. | | Deposit date: | 2019-04-02 | | Release date: | 2019-10-16 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.517 Å) | | Cite: | A Consensus Binding Motif for the PP4 Protein Phosphatase.
Mol.Cell, 76, 2019
|
|
5EZX
 
 | | CRYSTAL STRUCTURE OF BACE-1 IN COMPLEX WITH {(1R,2R)-2-[(R)-2-Amino-4-(4-difluoromethoxy-phenyl)-4,5-dihydro-oxazol-4-yl]-cyclopropyl}-(5-chloro-pyridin-3-yl)-methanone | | Descriptor: | Beta-secretase 1, DIMETHYL SULFOXIDE, SODIUM ION, ... | | Authors: | Banner, D, Benz, J, Stihle, M, Kuglstatter, A. | | Deposit date: | 2015-11-27 | | Release date: | 2016-02-24 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | A Real-World Perspective on Molecular Design.
J.Med.Chem., 59, 2016
|
|
7OOF
 
 | |
