2OP9
 
 | |
8FYU
 
 | | Crystal structure of the human CHIP-TPR domain in complex with a 10mer acetylated tau peptide | | Descriptor: | ACE-SER-SER-THR-GLY-SER-ILE-ASP-MET-VAL-ASP, E3 ubiquitin-protein ligase CHIP | | Authors: | Wucherer, K, Bohn, M.F, Basu, K, Nadel, C.M, Gestwicki, J.E, Craik, C.S. | | Deposit date: | 2023-01-26 | | Release date: | 2023-08-30 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.84839141 Å) | | Cite: | Phosphorylation of tau at a single residue inhibits binding to the E3 ubiquitin ligase, CHIP.
Nat Commun, 15, 2024
|
|
4W5C
 
 | | Crystal structure analysis of cruzain with three Fragments: 1 (N-(1H-benzimidazol-2-yl)-1,3-dimethyl-pyrazole-4-carboxamide), 6 (2-amino-4,6-difluorobenzothiazole) and 9 (N-(1H-benzimidazol-2-yl)-3-(4-fluorophenyl)-1H-pyrazole-4-carboxamide). | | Descriptor: | 4,6-difluoro-1,3-benzothiazol-2-amine, Cruzipain, N-(1H-benzimidazol-2-yl)-1,3-dimethyl-1H-pyrazole-4-carboxamide, ... | | Authors: | Tochowicz, A, McKerrow, J.H, Craik, C.S. | | Deposit date: | 2014-08-17 | | Release date: | 2015-04-08 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (3.27 Å) | | Cite: | Applying Fragments Based- Drug Design to identify multiple binding modes on cysteine protease.
To Be Published
|
|
1Z8G
 
 | | Crystal structure of the extracellular region of the transmembrane serine protease hepsin with covalently bound preferred substrate. | | Descriptor: | ACE-LYS-GLN-LEU-ARG-Chloromethylketone, Serine protease hepsin | | Authors: | Herter, S, Piper, D.E, Aaron, W, Gabriele, T, Cutler, G, Cao, P, Bhatt, A.S, Choe, Y, Craik, C.S, Walker, N, Meininger, D, Hoey, T, Austin, R.J. | | Deposit date: | 2005-03-30 | | Release date: | 2005-05-03 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Hepatocyte growth factor is a preferred in vitro substrate for human hepsin, a membrane-anchored serine protease implicated in prostate and ovarian cancers
Biochem.J., 390, 2005
|
|
4W5B
 
 | |
3SO3
 
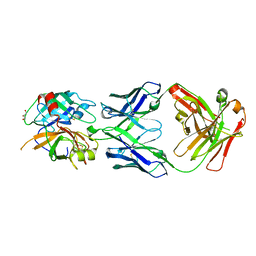 | | Structures of Fab-Protease Complexes Reveal a Highly Specific Non-Canonical Mechanism of Inhibition. | | Descriptor: | A11 FAB heavy chain, A11 FAB light chain, GLYCEROL, ... | | Authors: | Schneider, E.L, Farady, C.J, Egea, P.F, Goetz, D.H, Baharuddin, A, Craik, C.S. | | Deposit date: | 2011-06-29 | | Release date: | 2012-06-20 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | A reverse binding motif that contributes to specific protease inhibition by antibodies.
J.Mol.Biol., 415, 2012
|
|
5FMG
 
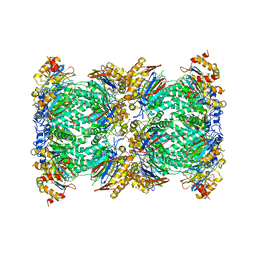 | | Structure and function based design of Plasmodium-selective proteasome inhibitors | | Descriptor: | (2S)-N-[(E,2S)-1-(1H-indol-3-yl)-4-methylsulfonyl-but-3-en-2-yl]-2-[[(2S)-3-(1H-indol-3-yl)-2-(2-morpholin-4-ylethanoylamino)propanoyl]amino]-4-methyl-pentanamide, BETA3 PROTEASOME SUBUNIT, PUTATIVE, ... | | Authors: | Li, H, O'Donoghue, A.J, van der Linden, W.A, Xie, S.C, Yoo, E, Foe, I.T, Tilley, L, Craik, C.S, da Fonseca, P.C.A, Bogyo, M. | | Deposit date: | 2015-11-04 | | Release date: | 2016-03-02 | | Last modified: | 2024-11-13 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Structure and Function Based Design of Plasmodium-Selective Proteasome Inhibitors
Nature, 530, 2016
|
|
3BN9
 
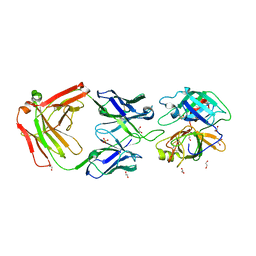 | | Crystal Structure of MT-SP1 in complex with Fab Inhibitor E2 | | Descriptor: | 1,2-ETHANEDIOL, E2 Fab Heavy Chain, E2 Fab Light Chain, ... | | Authors: | Farady, C.J, Schneider, E.L, Egea, P.F, Goetz, D.H, Craik, C.S. | | Deposit date: | 2007-12-13 | | Release date: | 2008-09-09 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.173 Å) | | Cite: | Structure of an Fab-protease complex reveals a highly specific non-canonical mechanism of inhibition
J.Mol.Biol., 380, 2008
|
|
2SAM
 
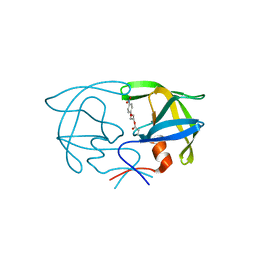 | | STRUCTURE OF THE PROTEASE FROM SIMIAN IMMUNODEFICIENCY VIRUS: COMPLEX WITH AN IRREVERSIBLE NON-PEPTIDE INHIBITOR | | Descriptor: | 3-(4-NITRO-PHENOXY)-PROPAN-1-OL, SIV PROTEASE | | Authors: | Rose, R.B, Rose, J.R, Salto, R, Craik, C.S, Stroud, R.M. | | Deposit date: | 1994-07-08 | | Release date: | 1994-10-15 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structure of the protease from simian immunodeficiency virus: complex with an irreversible nonpeptide inhibitor.
Biochemistry, 32, 1993
|
|
7TCZ
 
 | | Human cytomegalovirus protease mutant (C84A, C87A, C138A, C202A) in complex with inhibitor | | Descriptor: | Assemblin, [1-(2-oxopropyl)-4-phenyl-1H-1,2,3-triazol-5-yl]methyl benzylcarbamate | | Authors: | Hulce, K.R, Bohn, M, Ongpipattanakul, C, Jaishankar, P, Renslo, A.R, Craik, C.S. | | Deposit date: | 2021-12-29 | | Release date: | 2022-01-12 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.67 Å) | | Cite: | Inhibiting a dynamic viral protease by targeting a non-catalytic cysteine.
Cell Chem Biol, 29, 2022
|
|
8GCK
 
 | | Crystal structure of the human CHIP-TPR domain in complex with a 6mer acetylated tau peptide | | Descriptor: | ACE-SER-ILE-ASP-MET-VAL-ASP, E3 ubiquitin-protein ligase CHIP | | Authors: | Wucherer, K, Bohn, M.F, Basu, K, Nadel, C.M, Gestwicki, J.E, Craik, C.S. | | Deposit date: | 2023-03-02 | | Release date: | 2024-03-06 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.36823535 Å) | | Cite: | Intersecting PTMs regulate clearance of pathogenic tau by the ubiquitin ligase CHIP.
To Be Published
|
|
6BGJ
 
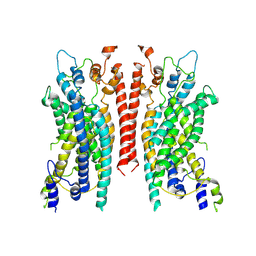 | | Cryo-EM structure of the TMEM16A calcium-activated chloride channel in LMNG | | Descriptor: | Anoctamin-1, CALCIUM ION | | Authors: | Dang, S, Feng, S, Tien, J, Peters, C.J, Bulkley, D, Lolicato, M, Zhao, J, Zuberbuhler, K, Ye, W, Qi, L, Chen, T, Craik, C.S, Jan, Y.N, Minor Jr, D.L, Cheng, Y, Jan, L.Y. | | Deposit date: | 2017-10-28 | | Release date: | 2017-12-27 | | Last modified: | 2025-06-04 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Cryo-EM structures of the TMEM16A calcium-activated chloride channel.
Nature, 552, 2017
|
|
6BGI
 
 | | Cryo-EM structure of the TMEM16A calcium-activated chloride channel in nanodisc | | Descriptor: | Anoctamin-1, CALCIUM ION | | Authors: | Dang, S, Feng, S, Tien, J, Peters, C.J, Bulkley, D, Lolicato, M, Zhao, J, Zuberbuhler, K, Ye, W, Qi, J, Chen, T, Craik, C.S, Jan, Y.N, Minor Jr, D.L, Cheng, Y, Jan, L.Y. | | Deposit date: | 2017-10-28 | | Release date: | 2017-12-27 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Cryo-EM structures of the TMEM16A calcium-activated chloride channel.
Nature, 552, 2017
|
|
8UDR
 
 | | Structure of the P1B7 antibody bound to the Sotorasib-modified KRas G12C peptide presented by the A*03:01 MHC I complex | | Descriptor: | AMG 510 (bound form), Beta-2-microglobulin, GTPase KRas, ... | | Authors: | Chan, L.M, Roweder, P.J, Craik, C.S, Verba, K.A. | | Deposit date: | 2023-09-28 | | Release date: | 2024-10-16 | | Last modified: | 2025-05-14 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Therapeutic Targeting and Structural Characterization of a Sotorasib-Modified KRAS G12C-MHC I Complex Demonstrate the Antitumor Efficacy of Hapten-Based Strategies.
Cancer Res., 85, 2025
|
|
6NSV
 
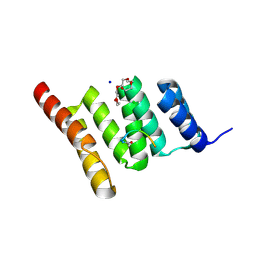 | | Crystal structure of the human CHIP TPR domain in complex with a 5mer acetylated optimized peptide | | Descriptor: | ACE-LEU-TRP-TRP-PRO-ASP, CHLORIDE ION, E3 ubiquitin-protein ligase CHIP, ... | | Authors: | Basu, K, Ravalin, M, Bohn, M.-F, Craik, C.S, Gestwicki, J.E. | | Deposit date: | 2019-01-25 | | Release date: | 2019-07-31 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.305 Å) | | Cite: | Specificity for latent C termini links the E3 ubiquitin ligase CHIP to caspases.
Nat.Chem.Biol., 15, 2019
|
|
5V5D
 
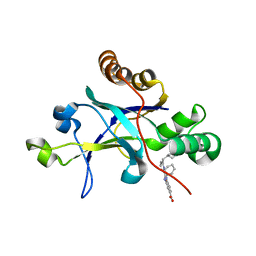 | | Room temperature (280K) crystal structure of Kaposi's sarcoma-associated herpesvirus protease in complex with allosteric inhibitor (compound 250) | | Descriptor: | 4-{[6-(cyclohexylmethyl)pyridine-2-carbonyl]amino}-3-(phenylamino)benzoic acid, ORF 17 | | Authors: | Thompson, M.C, Acker, T.M, Fraser, J.S, Craik, C.S. | | Deposit date: | 2017-03-14 | | Release date: | 2017-04-12 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.104 Å) | | Cite: | Allosteric Inhibitors, Crystallography, and Comparative Analysis Reveal Network of Coordinated Movement across Human Herpesvirus Proteases.
J. Am. Chem. Soc., 139, 2017
|
|
5UVP
 
 | |
5UR3
 
 | |
5UTN
 
 | |
5UV3
 
 | |
5UTE
 
 | |
5V5E
 
 | | Room temperature (280K) crystal structure of Kaposi's sarcoma-associated herpesvirus protease in complex with allosteric inhibitor (compound 733) | | Descriptor: | 4-{[6-(cyclohexylmethyl)pyridine-2-carbonyl]amino}-3-{[3-(trifluoromethoxy)phenyl]amino}benzoic acid, ORF 17 | | Authors: | Thompson, M.C, Acker, T.M, Fraser, J.S, Craik, C.S. | | Deposit date: | 2017-03-14 | | Release date: | 2017-04-12 | | Last modified: | 2024-12-25 | | Method: | X-RAY DIFFRACTION (2.299 Å) | | Cite: | Allosteric Inhibitors, Crystallography, and Comparative Analysis Reveal Network of Coordinated Movement across Human Herpesvirus Proteases.
J. Am. Chem. Soc., 139, 2017
|
|
1IFG
 
 | | CRYSTAL STRUCTURE OF A MONOMERIC FORM OF GENERAL PROTEASE INHIBITOR, ECOTIN IN ABSENCE OF A PROTEASE | | Descriptor: | ECOTIN | | Authors: | Eggers, C.T, Wang, S.X, Fletterick, R.J, Craik, C.S. | | Deposit date: | 2001-04-12 | | Release date: | 2001-05-18 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The role of ecotin dimerization in protease inhibition.
J.Mol.Biol., 308, 2001
|
|
1AZ5
 
 | |
1U9Q
 
 | | Crystal structure of cruzain bound to an alpha-ketoester | | Descriptor: | [1-(1-METHYL-4,5-DIOXO-PENT-2-ENYLCARBAMOYL)-2-PHENYL-ETHYL]-CARBAMIC ACID BENZYL ESTER, cruzipain | | Authors: | Lange, M, Weston, S.G, Cheng, H, Culliane, M, Fiorey, M.M, Grisostomi, C, Hardy, L.W, Hartstough, D.S, Pallai, P.V, Tilton, R.F, Baldino, C.M, Brinen, L.S, Engel, J.C, Choe, Y, Price, M.S, Craik, C.S. | | Deposit date: | 2004-08-10 | | Release date: | 2005-03-29 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Development of alpha-keto-based inhibitors of cruzain, a cysteine protease implicated in Chagas disease
Bioorg.Med.Chem., 13, 2005
|
|
