5KH5
 
 | |
5KH4
 
 | |
5KH2
 
 | |
6DXL
 
 | | Linked amidobenzimidazole STING agonist | | Descriptor: | 1,1'-(butane-1,4-diyl)bis{2-[(1-ethyl-3-methyl-1H-pyrazole-5-carbonyl)amino]-1H-benzimidazole-5-carboxamide}, CALCIUM ION, Stimulator of interferon protein | | Authors: | Concha, N.O. | | Deposit date: | 2018-06-29 | | Release date: | 2018-11-07 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Design of amidobenzimidazole STING receptor agonists with systemic activity.
Nature, 564, 2018
|
|
6DXG
 
 | | amidobenzimidazole (ABZI) STING agonists | | Descriptor: | 2-[(1-ethyl-3-methyl-1H-pyrazole-5-carbonyl)amino]-1-[(2R)-2-hydroxy-2-phenylethyl]-1H-benzimidazole-5-carboxamide, CALCIUM ION, Stimulator of interferon protein | | Authors: | Concha, N.O. | | Deposit date: | 2018-06-28 | | Release date: | 2018-11-07 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.905 Å) | | Cite: | Design of amidobenzimidazole STING receptor agonists with systemic activity.
Nature, 564, 2018
|
|
4L04
 
 | |
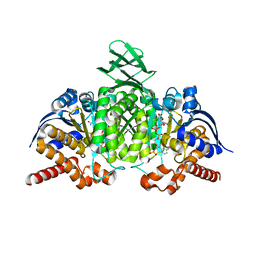
4L06
 
 | |
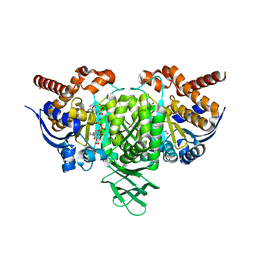
4L03
 
 | |
4KZO
 
 | |
3KKV
 
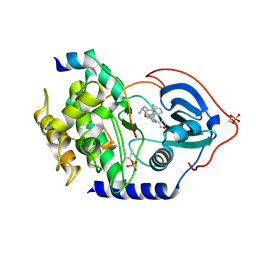 | | Structure of PKA with a protein Kinase B-selective inhibitor. | | Descriptor: | (2S)-1-{[6-furan-3-yl-5-(3-methyl-2H-indazol-5-yl)pyridin-3-yl]oxy}-3-(1H-indol-3-yl)propan-2-amine, PKI kinase inhibitor, cAMP-dependent protein kinase catalytic subunit alpha | | Authors: | Elkins, P.A, Concha, N.O. | | Deposit date: | 2009-11-06 | | Release date: | 2010-12-22 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structure of PKA with a protein Kinase B-selective inhibitor.
To be Published
|
|
1HLK
 
 | | METALLO-BETA-LACTAMASE FROM BACTEROIDES FRAGILIS IN COMPLEX WITH A TRICYCLIC INHIBITOR | | Descriptor: | 7,8-DIHYDROXY-1-METHOXY-3-METHYL-10-OXO-4,10-DIHYDRO-1H,3H-PYRANO[4,3-B]CHROMENE-9-CARBOXYLIC ACID, BETA-LACTAMASE, TYPE II, ... | | Authors: | Payne, D.J, Hueso-Rodriguez, J.A, Boyd, H, Concha, N.O, Janson, C.A, Gilpin, M, Bateson, J.H, Chever, C, Niconovich, N.L, Pearson, S, Rittenhouse, S, Tew, D, Diez, E, Perez, P, de la Fuente, J, Rees, M, Rivera-Sagredo, A. | | Deposit date: | 2000-12-01 | | Release date: | 2001-11-30 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Identification of a series of tricyclic natural products as potent broad spectrum inhibitors of metallo-beta-lactamases
Antimicrob.Agents Chemother., 46, 2002
|
|
8ULD
 
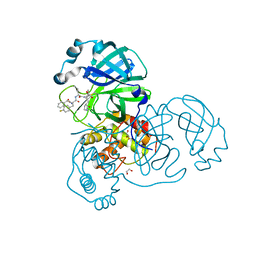 | | SARA CoV-2 3C-like protease in complex with GSK3487016A | | Descriptor: | 1,2-ETHANEDIOL, N-[(benzyloxy)carbonyl]-4-fluoro-L-phenylalanyl-N-{(2S,3R)-3-hydroxy-4-oxo-1-[(3S)-2-oxopyrrolidin-3-yl]-4-[(propan-2-yl)amino]butan-2-yl}-L-leucinamide, Replicase polyprotein 1a | | Authors: | Williams, S.P, Concha, N.O. | | Deposit date: | 2023-10-16 | | Release date: | 2024-02-14 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Exploration of the P1 residue in 3CL protease inhibitors leading to the discovery of a 2-tetrahydrofuran P1 replacement.
Bioorg.Med.Chem., 100, 2024
|
|
1KR3
 
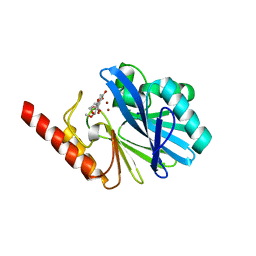 | | Crystal Structure of the Metallo beta-Lactamase from Bacteroides fragilis (CfiA) in Complex with the Tricyclic Inhibitor SB-236050. | | Descriptor: | 7,8-DIHYDROXY-1-METHOXY-3-METHYL-10-OXO-4,10-DIHYDRO-1H,3H-PYRANO[4,3-B]CHROMENE-9-CARBOXYLIC ACID, SODIUM ION, ZINC ION, ... | | Authors: | Payne, D.J, Hueso-Rodrguez, J.A, Boyd, H, Concha, N.O, Janson, C.A, Gilpin, M, Bateson, J.H, Cheever, C, Niconovich, N.L, Pearson, S, Rittenhouse, S, Tew, D, Dez, E, Prez, P, de la Fuente, J, Rees, M, Rivera-Sagredo, A. | | Deposit date: | 2002-01-08 | | Release date: | 2003-01-08 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Identification of a series of tricyclic natural products as potent broad-spectrum inhibitors of metallo-beta-lactamases
ANTIMICROB.AGENTS CHEMOTHER., 46, 2002
|
|
1A8A
 
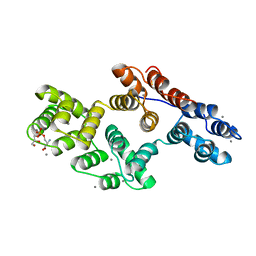 | | RAT ANNEXIN V COMPLEXED WITH GLYCEROPHOSPHOSERINE | | Descriptor: | ANNEXIN V, CALCIUM ION, L-ALPHA-GLYCEROPHOSPHORYLSERINE | | Authors: | Swairjo, M.A, Concha, N.O, Kaetzel, M.A, Dedman, J.R, Seaton, B.A. | | Deposit date: | 1998-03-23 | | Release date: | 1998-06-17 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Ca(2+)-bridging mechanism and phospholipid head group recognition in the membrane-binding protein annexin V.
Nat.Struct.Biol., 2, 1995
|
|
1A8B
 
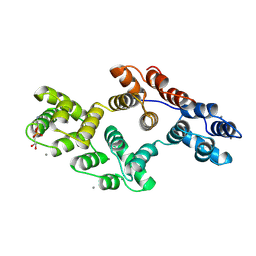 | | RAT ANNEXIN V COMPLEXED WITH GLYCEROPHOSPHOETHANOLAMINE | | Descriptor: | ANNEXIN V, CALCIUM ION, L-ALPHA-GLYCEROPHOSPHORYLETHANOLAMINE | | Authors: | Swairjo, M.A, Concha, N.O, Kaetzel, M.A, Dedman, J.R, Seaton, B.A. | | Deposit date: | 1998-03-23 | | Release date: | 1998-06-17 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Ca(2+)-bridging mechanism and phospholipid head group recognition in the membrane-binding protein annexin V.
Nat.Struct.Biol., 2, 1995
|
|
1NVQ
 
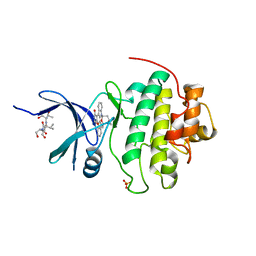 | | The Complex Structure Of Checkpoint Kinase Chk1/UCN-01 | | Descriptor: | 7-HYDROXYSTAUROSPORINE, Peptide ASVSA, SULFATE ION, ... | | Authors: | Zhao, B, Bower, M.J, McDevitt, P.J, Zhao, H, Davis, S.T, Johanson, K.O, Green, S.M, Concha, N.O, Zhou, B.B. | | Deposit date: | 2003-02-04 | | Release date: | 2003-04-08 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural Basis for Chk1 Inhibition by UCN-01
J.Biol.Chem., 277, 2002
|
|
1NVR
 
 | | The Complex Structure Of Checkpoint Kinase Chk1/Staurosporine | | Descriptor: | Peptide ASVSA, STAUROSPORINE, SULFATE ION, ... | | Authors: | Zhao, B, Bower, M.J, McDevitt, P.J, Zhao, H, Davis, S.T, Johanson, K.O, Green, S.M, Concha, N.O, Zhou, B.B. | | Deposit date: | 2003-02-04 | | Release date: | 2003-04-08 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural Basis for Chk1 Inhibition by UCN-01
J.Biol.Chem., 277, 2002
|
|
1NVS
 
 | | The Complex Structure Of Checkpoint Kinase Chk1/SB218078 | | Descriptor: | Peptide ASVSA, REL-(9R,12S)-9,10,11,12-TETRAHYDRO-9,12-EPOXY-1H-DIINDOLO[1,2,3-FG:3',2',1'-KL]PYRROLO[3,4-I][1,6]BENZODIAZOCINE-1,3(2H)-DIONE, SULFATE ION, ... | | Authors: | Zhao, B, Bower, M.J, McDevitt, P.J, Zhao, H, Davis, S.T, Johanson, K.O, Green, S.M, Concha, N.O, Zhou, B.B. | | Deposit date: | 2003-02-04 | | Release date: | 2003-04-08 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural Basis for Chk1 Inhibition by UCN-01
J.Biol.Chem., 277, 2002
|
|
7T0R
 
 | | Crystal structure of the anti-CD4 adnectin 6940_B01 as a complex with the extracellular domains of CD4 and ibalizumab fAb | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Adnectin 6940_B01, Ibalizumab Heavy Chain, ... | | Authors: | Williams, S.P, Concha, N.O, Wensel, D.L, Hong, X. | | Deposit date: | 2021-11-30 | | Release date: | 2022-01-12 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (3.65 Å) | | Cite: | Novel Bent Conformation of CD4 Induced by HIV-1 Inhibitor Indirectly Prevents Productive Viral Attachment.
J.Mol.Biol., 434, 2021
|
|
