4Y1H
 
 | | Crystal structure of K33 linked tri-Ubiquitin | | Descriptor: | 1,2-ETHANEDIOL, Ubiquitin-40S ribosomal protein S27a | | Authors: | Kristariyanto, Y.A, Abdul Rehman, S.A, Choi, S.Y, Ritorto, S, Campbell, D.G, Morrice, N.A, Toth, R, Kulathu, Y. | | Deposit date: | 2015-02-07 | | Release date: | 2015-03-18 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Assembly and structure of Lys33-linked polyubiquitin reveals distinct conformations.
Biochem.J., 467, 2015
|
|
5TZL
 
 | | Structure of transthyretin in complex with the kinetic stabilizer 201 | | Descriptor: | 4-(7-chloro-1,3-benzoxazol-2-yl)-2,6-diiodophenol, Transthyretin | | Authors: | Connelly, S, Mortenson, D.E, Choi, S, Wilson, I.A, Powers, E.T, Kelly, J.W, Johnson, S.M. | | Deposit date: | 2016-11-21 | | Release date: | 2017-06-28 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Semi-quantitative models for identifying potent and selective transthyretin amyloidogenesis inhibitors.
Bioorg. Med. Chem. Lett., 27, 2017
|
|
4XYZ
 
 | | Crystal structure of K33 linked di-Ubiquitin | | Descriptor: | 1,2-ETHANEDIOL, ACETATE ION, IODIDE ION, ... | | Authors: | Kristariyanto, Y.A, Abdul Rehman, S.A, Choi, S.Y, Ritorto, S, Campbell, D.G, Morrice, N.A, Toth, R, Kulathu, Y. | | Deposit date: | 2015-02-03 | | Release date: | 2015-03-18 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Assembly and structure of Lys33-linked polyubiquitin reveals distinct conformations.
Biochem.J., 467, 2015
|
|
8GSI
 
 | | Structure of the cobolimab Fab | | Descriptor: | heavy chain, light chain, nanobody | | Authors: | Heo, Y.S, Choi, S.B. | | Deposit date: | 2022-09-06 | | Release date: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.02 Å) | | Cite: | Crystal structure of the Fab fragment of cobolimab, an investigational antibody targeting Tim-3 for immune-oncology.
To Be Published
|
|
4ZJ1
 
 | | Crystal Structure of p-acrylamido-phenylalanine modified TEM1 beta-lactamase from Escherichia coli : V216AcrF mutant | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, Beta-lactamase TEM, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Xiao, H, Nasertorabi, F, Choi, S, Han, G.W, Reed, S.A, Stevens, R.C, Schultz, P.G. | | Deposit date: | 2015-04-28 | | Release date: | 2015-05-20 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.54 Å) | | Cite: | Exploring the potential impact of an expanded genetic code on protein function.
Proc.Natl.Acad.Sci.USA, 112, 2015
|
|
4ZJ3
 
 | | Crystal structure of cephalexin bound acyl-enzyme intermediate of Val216AcrF mutant TEM1 beta-lactamase from Escherichia coli: E166N and V216AcrF mutant. | | Descriptor: | Beta-lactamase TEM | | Authors: | Xiao, H, Nasertorabi, F, Choi, S, Han, G.W, Reed, S.A, Stevens, R.C, Schultz, P.G. | | Deposit date: | 2015-04-29 | | Release date: | 2015-05-20 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Exploring the potential impact of an expanded genetic code on protein function.
Proc.Natl.Acad.Sci.USA, 112, 2015
|
|
3NJ4
 
 | | Fluoro-neplanocin A in Human S-Adenosylhomocysteine Hydrolase | | Descriptor: | (4S,5S)-4-(6-amino-9H-purin-9-yl)-3-fluoro-5-hydroxy-2-(hydroxymethyl)cyclopent-2-en-1-one, Adenosylhomocysteinase, NICOTINAMIDE-ADENINE-DINUCLEOTIDE | | Authors: | Jeong, L.S, Lee, K.M, Hwang, K.Y, Choi, S, Heo, Y.S. | | Deposit date: | 2010-06-17 | | Release date: | 2011-05-04 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | X-ray crystal structure and binding mode analysis of human S-adenosylhomocysteine hydrolase complexed with novel mechanism-based inhibitors, haloneplanocin A analogues.
J.Med.Chem., 54, 2011
|
|
4ZJ2
 
 | | Crystal Structure of p-acrylamido-phenylalanine modified TEM1 beta-lactamase from Escherichia coli :E166N mutant | | Descriptor: | Beta-lactamase TEM | | Authors: | Xiao, H, Nasertorabi, F, Choi, S, Han, G.W, Reed, S.A, Stevens, C.S, Schultz, P.G. | | Deposit date: | 2015-04-28 | | Release date: | 2015-05-20 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Exploring the potential impact of an expanded genetic code on protein function.
Proc.Natl.Acad.Sci.USA, 112, 2015
|
|
7D5Y
 
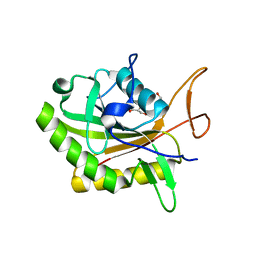 | |
4BGG
 
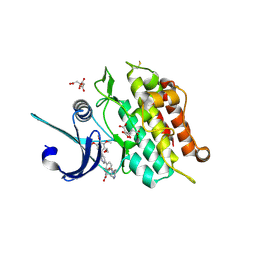 | | Crystal structure of the ACVR1 kinase in complex with LDN-213844 | | Descriptor: | 1,2-ETHANEDIOL, 1-{4-[5-(3,4,5-trimethoxyphenyl)pyridin-3-yl]phenyl}piperazine, ACTIVIN RECEPTOR TYPE-1, ... | | Authors: | Sanvitale, C, Canning, P, Cooper, C, Wang, Y, Mohedas, A.H, Choi, S, Yu, P.B, Cuny, G.D, Nowak, R, Coutandin, D, Vollmar, M, von Delft, F, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Bullock, A, Structural Genomics Consortium (SGC) | | Deposit date: | 2013-03-26 | | Release date: | 2013-04-10 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.56 Å) | | Cite: | Structure-Activity Relationship of 3,5-Diaryl-2-Aminopyridine Alk2 Inhibitors Reveals Unaltered Binding Affinity for Fibrodysplasia Ossificans Progressiva Causing Mutants.
J.Med.Chem., 57, 2014
|
|
3B1F
 
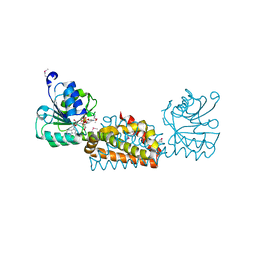 | | Crystal structure of prephenate dehydrogenase from Streptococcus mutans | | Descriptor: | NICOTINAMIDE-ADENINE-DINUCLEOTIDE, Putative prephenate dehydrogenase | | Authors: | Ku, H.K, Do, N.H, Song, J.S, Choi, S, Shin, M.H, Kim, K.J, Lee, S.J. | | Deposit date: | 2011-07-02 | | Release date: | 2011-10-26 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structure of prephenate dehydrogenase from Streptococcus mutans.
Int.J.Biol.Macromol., 49, 2011
|
|
4KY2
 
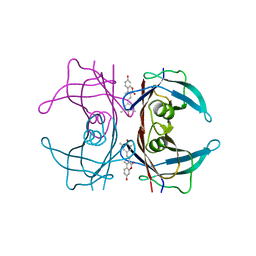 | | Transthyretin in complex with the fluorescent folding sensor (E)-7-hydroxy-3-(4-hydroxy-3,5-dimethylstyryl)-4-methyl-2H-chromen-2-one | | Descriptor: | 7-hydroxy-3-[(E)-2-(4-hydroxy-3,5-dimethylphenyl)ethenyl]-4-methyl-2H-chromen-2-one, Transthyretin | | Authors: | Connelly, S, Wilson, I.A, Choi, S. | | Deposit date: | 2013-05-28 | | Release date: | 2013-08-28 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.13 Å) | | Cite: | Bifunctional coumarin derivatives that inhibit transthyretin amyloidogenesis and serve as fluorescent transthyretin folding sensors.
Chem.Commun.(Camb.), 49, 2013
|
|
4LUB
 
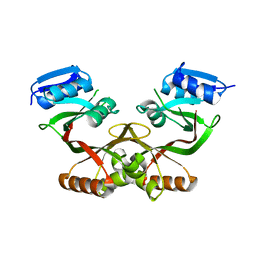 | |
3E4A
 
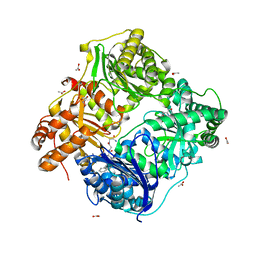 | | Human IDE-inhibitor complex at 2.6 angstrom resolution | | Descriptor: | 1,4-DIETHYLENE DIOXIDE, ACETIC ACID, HYDROXAMATE PEPTIDE II1, ... | | Authors: | Malito, E, Leissring, M.A, Choi, S, Cuny, G.D, Tang, W.J. | | Deposit date: | 2008-08-11 | | Release date: | 2009-05-19 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Designed inhibitors of insulin-degrading enzyme regulate the catabolism and activity of insulin.
Plos One, 5, 2010
|
|
7V5C
 
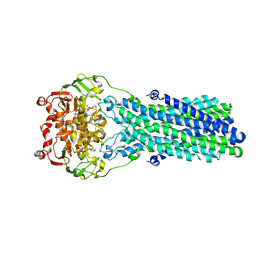 | | Cryo-EM structure of the mouse ABCB9 (ADP.BeF3-bound) | | Descriptor: | ABC-type oligopeptide transporter ABCB9, ADENOSINE-5'-DIPHOSPHATE, BERYLLIUM TRIFLUORIDE ION, ... | | Authors: | Park, J.G, Kim, S, Jang, E, Choi, S.H, Han, H, Kim, J.W, Ju, S, Min, D.S, Jin, M.S. | | Deposit date: | 2021-08-17 | | Release date: | 2022-10-19 | | Last modified: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | The lysosomal transporter TAPL has a dual role as peptide translocator and phosphatidylserine floppase.
Nat Commun, 13, 2022
|
|
7V5D
 
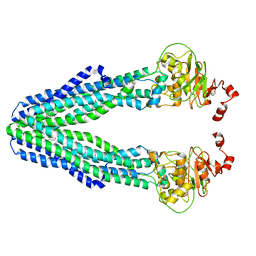 | | Cryo-EM structure of the mouse ABCB9 (PG-bound) | | Descriptor: | (1S)-2-{[{[(2R)-2,3-DIHYDROXYPROPYL]OXY}(HYDROXY)PHOSPHORYL]OXY}-1-[(PALMITOYLOXY)METHYL]ETHYL STEARATE, ABC-type oligopeptide transporter ABCB9 | | Authors: | Park, J.G, Kim, S, Jang, E, Choi, S.H, Han, H, Ju, S, Kim, J.W, Min, D.S, Jin, M.S. | | Deposit date: | 2021-08-17 | | Release date: | 2022-10-19 | | Last modified: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | The lysosomal transporter TAPL has a dual role as peptide translocator and phosphatidylserine floppase.
Nat Commun, 13, 2022
|
|
7VFI
 
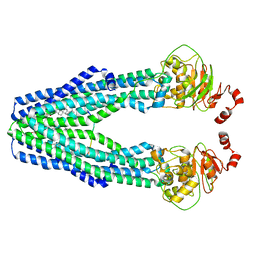 | | Cryo-EM structure of the mouse TAPL (9mer-peptide bound) | | Descriptor: | ABC-type oligopeptide transporter ABCB9, ARG-ARG-TYR-GLN-LYS-SER-THR-GLU-LEU, CHOLESTEROL HEMISUCCINATE | | Authors: | Park, J.G, Kim, S, Jang, E, Choi, S.H, Han, H, Kim, J.W, Ju, S, Min, D.S, Jin, M.S. | | Deposit date: | 2021-09-13 | | Release date: | 2022-10-19 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (3.98 Å) | | Cite: | The lysosomal transporter TAPL has a dual role as peptide translocator and phosphatidylserine floppase.
Nat Commun, 13, 2022
|
|
3OYY
 
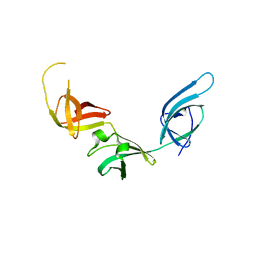 | |
3TVZ
 
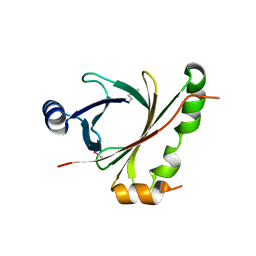 | | Structure of Bacillus subtilis HmoB | | Descriptor: | Putative uncharacterized protein yhgC | | Authors: | Choe, J, Choi, S, Park, S. | | Deposit date: | 2011-09-21 | | Release date: | 2012-07-11 | | Last modified: | 2019-10-09 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Bacillus subtilis HmoB is a heme oxygenase with a novel structure.
Bmb Rep, 45, 2012
|
|
6II6
 
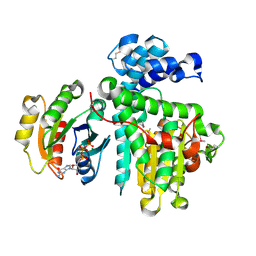 | | Crystal structure of the Makes Caterpillars Floppy (MCF)-Like effector of Vibrio vulnificus MO6-24/O in complex with a human ADP-ribosylation factor 3 (ARF3) | | Descriptor: | ADP-ribosylation factor 3, GUANOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Lee, Y, Kim, B.S, Choi, S, Lee, E.Y, Park, S, Hwang, J, Kwon, Y, Hyun, J, Lee, C, Eom, S.H, Kim, M.H. | | Deposit date: | 2018-10-03 | | Release date: | 2019-08-07 | | Last modified: | 2020-02-19 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Makes caterpillars floppy-like effector-containing MARTX toxins require host ADP-ribosylation factor (ARF) proteins for systemic pathogenicity.
Proc.Natl.Acad.Sci.USA, 116, 2019
|
|
6IM3
 
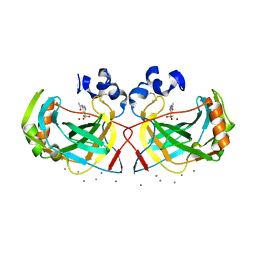 | | Crystal structure of a highly thermostable carbonic anhydrase from Persephonella marina EX-H1 | | Descriptor: | 5-ACETAMIDO-1,3,4-THIADIAZOLE-2-SULFONAMIDE, CALCIUM ION, Carbonic anhydrase (Carbonate dehydratase), ... | | Authors: | Jin, M.S, Kim, S, Sung, J, Yeon, J, Choi, S.H. | | Deposit date: | 2018-10-22 | | Release date: | 2019-07-17 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal Structure of a Highly Thermostable alpha-Carbonic Anhydrase from Persephonella marina EX-H1.
Mol.Cells, 42, 2019
|
|
6II2
 
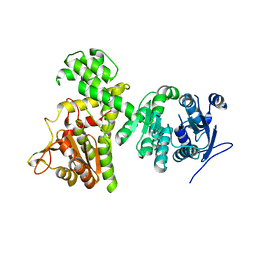 | | Crystal structure of alpha-beta hydrolase (ABH) and Makes Caterpillars Floppy (MCF)-Like effectors of Vibrio vulnificus MO6-24/O | | Descriptor: | Putative RTX-toxin | | Authors: | Lee, Y, Kim, B.S, Choi, S, Lee, E.Y, Park, S, Hwang, J, Kwon, Y, Hyung, J, Lee, C, Eom, S.H, Kim, M.H. | | Deposit date: | 2018-10-03 | | Release date: | 2019-08-07 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Makes caterpillars floppy-like effector-containing MARTX toxins require host ADP-ribosylation factor (ARF) proteins for systemic pathogenicity.
Proc.Natl.Acad.Sci.USA, 116, 2019
|
|
6II0
 
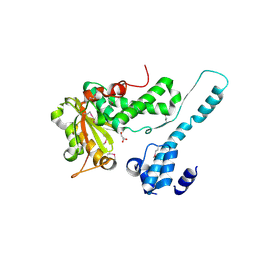 | | Crystal structure of the Makes Caterpillars Floppy (MCF)-Like effector of Vibrio vulnificus MO6-24/O | | Descriptor: | GLYCEROL, Putative RTX-toxin | | Authors: | Lee, Y, Kim, B.S, Choi, S, Lee, E.Y, Park, S, Hwang, J, Kwon, Y, Hyun, J, Lee, C, Eom, S.H, Kim, M.H. | | Deposit date: | 2018-10-03 | | Release date: | 2019-08-07 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.36 Å) | | Cite: | Makes caterpillars floppy-like effector-containing MARTX toxins require host ADP-ribosylation factor (ARF) proteins for systemic pathogenicity.
Proc.Natl.Acad.Sci.USA, 116, 2019
|
|
6IM1
 
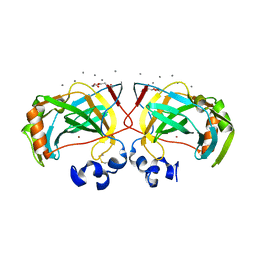 | | Crystal structure of a highly thermostable carbonic anhydrase from Persephonella marina EX-H1 | | Descriptor: | CALCIUM ION, Carbonic anhydrase (Carbonate dehydratase), TETRAETHYLENE GLYCOL, ... | | Authors: | Jin, M.S, Kim, S, Sung, J, Yeon, J, Choi, S.H. | | Deposit date: | 2018-10-21 | | Release date: | 2019-07-17 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal Structure of a Highly Thermostable alpha-Carbonic Anhydrase from Persephonella marina EX-H1.
Mol.Cells, 42, 2019
|
|
6IM0
 
 | | Crystal structure of a highly thermostable carbonic anhydrase from Persephonella marina EX-H1 | | Descriptor: | BICARBONATE ION, CALCIUM ION, Carbonic anhydrase (Carbonate dehydratase), ... | | Authors: | Jin, M.S, Kim, S, Sung, J, Yeon, J, Choi, S.H. | | Deposit date: | 2018-10-21 | | Release date: | 2019-07-17 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Crystal Structure of a Highly Thermostable alpha-Carbonic Anhydrase from Persephonella marina EX-H1.
Mol.Cells, 42, 2019
|
|
