6XJT
 
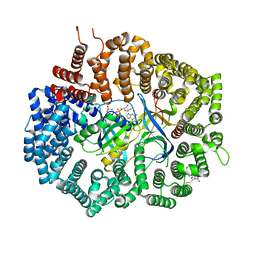 | | Crystal Structure of KPT-8602 bound to CRM1 (537-DLTVK-541 to GLCEQ) | | Descriptor: | (2R)-3-{3-[3,5-bis(trifluoromethyl)phenyl]-1H-1,2,4-triazol-1-yl}-2-(pyrimidin-5-yl)propanamide, Exportin-1, GTP-binding nuclear protein Ran, ... | | Authors: | Baumhardt, J.M, Chook, Y.M. | | Deposit date: | 2020-06-24 | | Release date: | 2021-01-27 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.406 Å) | | Cite: | Recurrent XPO1 mutations alter pathogenesis of chronic lymphocytic leukemia.
J Hematol Oncol, 14, 2021
|
|
6XJU
 
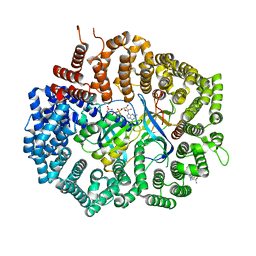 | | Crystal Structure of KPT-8602 bound to CRM1 (E582K, 537-DLTVK-541 to GLCEQ) | | Descriptor: | (2R)-3-{3-[3,5-bis(trifluoromethyl)phenyl]-1H-1,2,4-triazol-1-yl}-2-(pyrimidin-5-yl)propanamide, Exportin-1, GTP-binding nuclear protein Ran, ... | | Authors: | Baumhardt, J.M, Chook, Y.M. | | Deposit date: | 2020-06-24 | | Release date: | 2021-01-27 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.193 Å) | | Cite: | Recurrent XPO1 mutations alter pathogenesis of chronic lymphocytic leukemia.
J Hematol Oncol, 14, 2021
|
|
6XJS
 
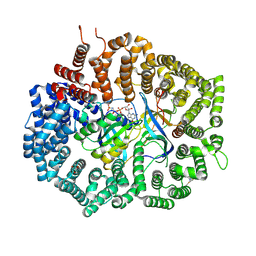 | |
3GB8
 
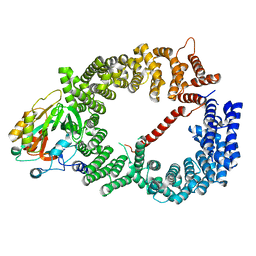 | | Crystal structure of CRM1/Snurportin-1 complex | | Descriptor: | Exportin-1, Snurportin-1 | | Authors: | Dong, X, Biswas, A, Suel, K.E, Jackson, L.K, Martinez, R, Gu, H, Chook, Y.M. | | Deposit date: | 2009-02-19 | | Release date: | 2009-03-31 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structural basis for leucine-rich nuclear export signal recognition by CRM1.
Nature, 458, 2009
|
|
4A4D
 
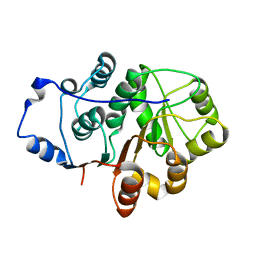 | | Crystal structure of the N-terminal domain of the Human DEAD-BOX RNA helicase DDX5 (P68) | | Descriptor: | PROBABLE ATP-DEPENDENT RNA HELICASE DDX5 | | Authors: | Dutta, S, Choi, Y.W, Kotaka, M, Fielding, B.C, Tan, Y.J. | | Deposit date: | 2011-10-11 | | Release date: | 2012-08-08 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | The Variable N-Terminal Region of Ddx5 Contains Structural Elements and Auto-Inhibits its Interaction with Ns5B of Hepatitis C Virus.
Biochem.J., 446, 2012
|
|
8SGH
 
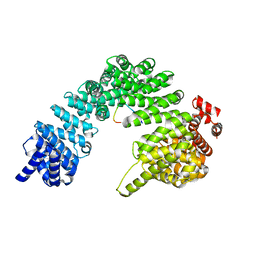 | |
2B1G
 
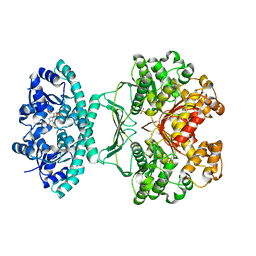 | | Crystal structures of transition state analogue inhibitors of inosine monophosphate cyclohydrolase | | Descriptor: | 7-(3,4-DIHYDROXY-5R-HYDROXYMETHYLTETRAHYDROFURAN-2-YL)-2,2-DIOXO-1,2R,3R,7-TETRAHYDRO-2L6-IMIDAZO[4,5-C][1,2,6]THIADIAZIN-4S-ONE, Bifunctional purine biosynthesis protein PURH, PHOSPHATE ION, ... | | Authors: | Xu, L, Chong, Y, Hwang, I, D'Onofrio, A, Amore, K, Beardsley, G.P, Li, C, Olson, A.J, Boger, D.L, Wilson, I.A. | | Deposit date: | 2005-09-15 | | Release date: | 2006-11-21 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structure-based Design, Synthesis, Evaluation, and Crystal Structures of Transition State Analogue Inhibitors of Inosine Monophosphate Cyclohydrolase.
J.Biol.Chem., 282, 2007
|
|
1LB4
 
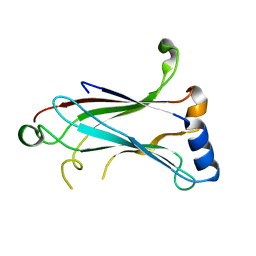 | | TRAF6 apo structure | | Descriptor: | TNF receptor-associated factor 6 | | Authors: | Ye, H, Arron, J.R, Lamothe, B, Cirilli, M, Kobayashi, T, Shevde, N.K, Segal, D, Dzivenu, O, Vologodskaia, M, Yim, M, Du, K, Singh, S, Pike, J.W, Darnay, B.G, Choi, Y, Wu, H. | | Deposit date: | 2002-04-02 | | Release date: | 2002-07-31 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Distinct molecular mechanism for initiating TRAF6 signalling.
Nature, 418, 2002
|
|
1LB5
 
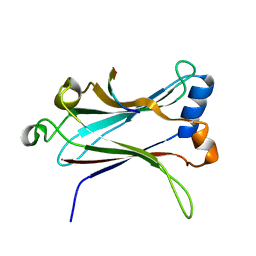 | | TRAF6-RANK Complex | | Descriptor: | TNF receptor-associated factor 6, receptor activator of nuclear factor-kappa B | | Authors: | Ye, H, Arron, J.R, Lamothe, B, Cirilli, M, Kobayashi, T, Shevde, N.K, Segal, D, Dzivenu, O, Vologodskaia, M, Yim, M, Du, K, Singh, S, Pike, J.W, Darnay, B.G, Choi, Y, Wu, H. | | Deposit date: | 2002-04-02 | | Release date: | 2002-07-31 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Distinct molecular mechanism for initiating TRAF6 signalling.
Nature, 418, 2002
|
|
1LB6
 
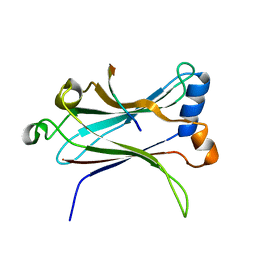 | | TRAF6-CD40 Complex | | Descriptor: | CD40 antigen, TNF receptor-associated factor 6 | | Authors: | Ye, H, Arron, J.R, Lamothe, B, Cirilli, M, Kobayashi, T, Shevde, N.K, Segal, D, Dzivenu, O, Vologodskaia, M, Yim, M, Du, K, Singh, S, Pike, J.W, Darnay, B.G, Choi, Y, Wu, H. | | Deposit date: | 2002-04-02 | | Release date: | 2002-07-31 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Distinct molecular mechanism for initiating TRAF6 signalling.
Nature, 418, 2002
|
|
2B1I
 
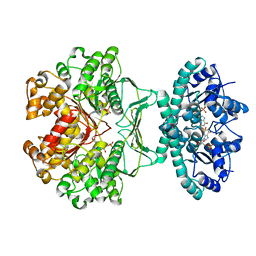 | | crystal structures of transition state analogue inhibitors of inosine monophosphate cyclohydrolase | | Descriptor: | Bifunctional purine biosynthesis protein PURH, POTASSIUM ION, [3,4-DIHYDROXY-5R-(2,2,4-TRIOXO-1,2R,3S,4R-TETRAHYDRO-2L6-IMIDAZO[4,5-C][1,2,6]THIADIAZIN-7-YL)TETRAHYDROFURAN-2-YL]METHYL DIHYDROGEN PHOSPHATE | | Authors: | Xu, L, Chong, Y, Hwang, I, D'Onofrio, A, Amore, K, Beardsley, G.P, Li, C, Olson, A.J, Boger, D.L, Wilson, I.A. | | Deposit date: | 2005-09-15 | | Release date: | 2006-11-21 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.02 Å) | | Cite: | Structure-based Design, Synthesis, Evaluation, and Crystal Structures of Transition State Analogue Inhibitors of Inosine Monophosphate Cyclohydrolase.
J.Biol.Chem., 282, 2007
|
|
7E60
 
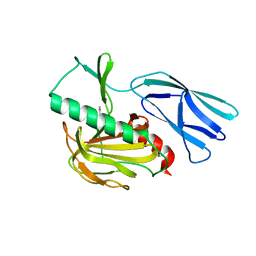 | | The crystal structure of peptidoglycan peptidase in complex with inhibitor 1 | | Descriptor: | (2~{R},6~{S})-2,6-diacetamido-7-[[(2~{R})-1-(oxidanylamino)-1-oxidanylidene-propan-2-yl]amino]-7-oxidanylidene-heptanoic acid, Peptidase M23, ZINC ION | | Authors: | Min, K, Yoon, H.J, Choi, Y, Lee, H.H. | | Deposit date: | 2021-02-21 | | Release date: | 2022-02-23 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.24 Å) | | Cite: | Structure-based inhibitor design for reshaping bacterial morphology
Commun Biol, 5, 2022
|
|
7E61
 
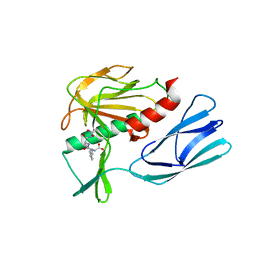 | | The crystal structure of peptidoglycan peptidase in complex with inhibitor 2 | | Descriptor: | 2-[[(3S)-3-acetamido-4-[[(2R)-1-(oxidanylamino)-1-oxidanylidene-propan-2-yl]amino]-4-oxidanylidene-butyl]-(phenylmethyl)amino]ethanoic acid, Peptidase M23, ZINC ION | | Authors: | Min, K.J, Yoon, H.J, Choi, Y, Lee, H.H. | | Deposit date: | 2021-02-21 | | Release date: | 2022-02-23 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structure-based inhibitor design for reshaping bacterial morphology
Commun Biol, 5, 2022
|
|
1U9Q
 
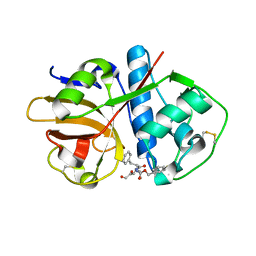 | | Crystal structure of cruzain bound to an alpha-ketoester | | Descriptor: | [1-(1-METHYL-4,5-DIOXO-PENT-2-ENYLCARBAMOYL)-2-PHENYL-ETHYL]-CARBAMIC ACID BENZYL ESTER, cruzipain | | Authors: | Lange, M, Weston, S.G, Cheng, H, Culliane, M, Fiorey, M.M, Grisostomi, C, Hardy, L.W, Hartstough, D.S, Pallai, P.V, Tilton, R.F, Baldino, C.M, Brinen, L.S, Engel, J.C, Choe, Y, Price, M.S, Craik, C.S. | | Deposit date: | 2004-08-10 | | Release date: | 2005-03-29 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Development of alpha-keto-based inhibitors of cruzain, a cysteine protease implicated in Chagas disease
Bioorg.Med.Chem., 13, 2005
|
|
8F19
 
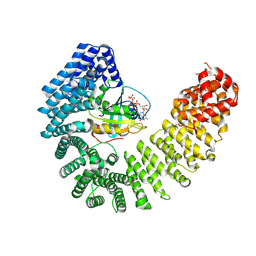 | | Cryo-EM structure of Kap114 bound to Gsp1 (RanGTP) | | Descriptor: | GTP-binding nuclear protein GSP1/CNR1, GUANOSINE-5'-TRIPHOSPHATE, Importin subunit beta-5, ... | | Authors: | Jiou, J, Chook, Y.M. | | Deposit date: | 2022-11-04 | | Release date: | 2023-07-26 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (3.49 Å) | | Cite: | Mechanism of RanGTP priming H2A-H2B release from Kap114 in an atypical RanGTP•Kap114•H2A-H2B complex.
Proc.Natl.Acad.Sci.USA, 120, 2023
|
|
8F1E
 
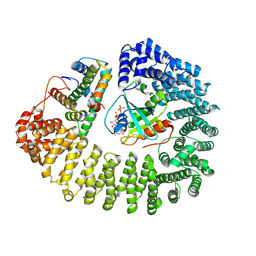 | | Cryo-EM structure of Kap114 bound to Gsp1 (RanGTP) and H2A-H2B | | Descriptor: | GTP-binding nuclear protein GSP1/CNR1, GUANOSINE-5'-TRIPHOSPHATE, Histone H2A.2, ... | | Authors: | Jiou, J, Chook, Y.M. | | Deposit date: | 2022-11-04 | | Release date: | 2023-07-26 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (3.28 Å) | | Cite: | Mechanism of RanGTP priming H2A-H2B release from Kap114 in an atypical RanGTP•Kap114•H2A-H2B complex.
Proc.Natl.Acad.Sci.USA, 120, 2023
|
|
8F0X
 
 | | Cryo-EM structure of Kap114 bound to H2A-H2B | | Descriptor: | Histone H2A.2, Histone H2B.2, Importin subunit beta-5 | | Authors: | Jiou, J, Chook, Y.M. | | Deposit date: | 2022-11-04 | | Release date: | 2023-07-26 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (3.21 Å) | | Cite: | Mechanism of RanGTP priming H2A-H2B release from Kap114 in an atypical RanGTP•Kap114•H2A-H2B complex.
Proc.Natl.Acad.Sci.USA, 120, 2023
|
|
8HST
 
 | |
8HSV
 
 | | The structure of rat beta-arrestin1 in complex with a rat Mdm2 peptide | | Descriptor: | Beta-arrestin-1, SULFATE ION, peptide from E3 ubiquitin-protein ligase Mdm2 | | Authors: | Yun, Y, Yoon, H.J, Choi, Y, Lee, H.H. | | Deposit date: | 2022-12-20 | | Release date: | 2023-07-19 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | GPCR targeting of E3 ubiquitin ligase MDM2 by inactive beta-arrestin.
Proc.Natl.Acad.Sci.USA, 120, 2023
|
|
8F7A
 
 | | Cryo-EM structure of Importin-9 bound to RanGTP | | Descriptor: | GTP-binding nuclear protein GSP1/CNR1, GUANOSINE-5'-TRIPHOSPHATE, Importin-9, ... | | Authors: | Bernardes, N.E, Chook, Y.M. | | Deposit date: | 2022-11-18 | | Release date: | 2023-07-26 | | Last modified: | 2024-05-22 | | Method: | ELECTRON MICROSCOPY (3.78 Å) | | Cite: | Mechanism of RanGTP priming H2A-H2B release from Kap114 in an atypical RanGTP•Kap114•H2A-H2B complex.
Proc.Natl.Acad.Sci.USA, 120, 2023
|
|
7CYL
 
 | |
7UNK
 
 | | Structure of Importin-4 bound to the H3-H4-ASF1 histone-histone chaperone complex | | Descriptor: | Histone H3, Histone H4, Histone chaperone, ... | | Authors: | Bernardes, N.E, Chook, Y.M, Fung, H.Y.J, Chen, Z, Li, Y. | | Deposit date: | 2022-04-11 | | Release date: | 2022-09-21 | | Last modified: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (3.45 Å) | | Cite: | Structure of IMPORTIN-4 bound to the H3-H4-ASF1 histone-histone chaperone complex.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
2IU3
 
 | | Crystal structures of transition state analogue inhibitors of inosine monophosphate cyclohydrolase | | Descriptor: | 1,5-DIHYDROIMIDAZO[4,5-C][1,2,6]THIADIAZIN-4(3H)-ONE 2,2-DIOXIDE, BIFUNCTIONAL PURINE BIOSYNTHESIS PROTEIN PURH, POTASSIUM ION | | Authors: | Xu, L, Chong, Y, Hwang, I, D'Onofrio, A, Amore, K, Beardsley, G.P, Li, C, Olson, A.J, Boger, D.L, Wilson, I.A. | | Deposit date: | 2006-05-27 | | Release date: | 2007-02-20 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structure-based design, synthesis, evaluation, and crystal structures of transition state analogue inhibitors of inosine monophosphate cyclohydrolase.
J. Biol. Chem., 282, 2007
|
|
2IU0
 
 | | crystal structures of transition state analogue inhibitors of inosine monophosphate cyclohydrolase | | Descriptor: | 1,5-DIHYDROIMIDAZO[4,5-C][1,2,6]THIADIAZIN-4(3H)-ONE 2,2-DIOXIDE, BIFUNCTIONAL PURINE BIOSYNTHESIS PROTEIN PURH, POTASSIUM ION | | Authors: | Xu, L, Chong, Y, Hwang, I, Onofrio, A.D, Amore, K, Beardsley, G.P, Li, C, Olson, A.J, Boger, D.L, Wilson, I.A. | | Deposit date: | 2006-05-26 | | Release date: | 2007-02-20 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.53 Å) | | Cite: | Structure-Based Design, Synthesis, Evaluation, and Crystal Structures of Transition State Analogue Inhibitors of Inosine Monophosphate Cyclohydrolase
J.Biol.Chem., 282, 2007
|
|
2OT8
 
 | |
