7EWR
 
 | | Cryo-EM structure of human GPR158 in complex with RGS7-Gbeta5 in a 2:2:2 ratio | | Descriptor: | Guanine nucleotide-binding protein subunit beta-5, Probable G-protein coupled receptor 158, Regulator of G-protein signaling 7 | | Authors: | Kim, Y, Jeong, E, Jeong, J, Cho, Y. | | Deposit date: | 2021-05-26 | | Release date: | 2021-12-01 | | Last modified: | 2022-02-16 | | Method: | ELECTRON MICROSCOPY (4.7 Å) | | Cite: | Structure of the class C orphan GPCR GPR158 in complex with RGS7-G beta 5.
Nat Commun, 12, 2021
|
|
7CAM
 
 | | SARS-CoV-2 main protease (Mpro) apo structure (space group P212121) | | Descriptor: | 3C-like proteinase | | Authors: | Wang, Y.C, Yang, C.S, Hou, M.H, Tsai, C.L, Chou, Y.Z, Chen, Y. | | Deposit date: | 2020-06-09 | | Release date: | 2021-05-05 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | Structural basis of SARS-CoV-2 main protease inhibition by a broad-spectrum anti-coronaviral drug.
Am J Cancer Res, 10, 2020
|
|
7CB7
 
 | | 1.7A resolution structure of SARS-CoV-2 main protease (Mpro) in complex with broad-spectrum coronavirus protease inhibitor GC376 | | Descriptor: | (1R,2S)-2-({N-[(benzyloxy)carbonyl]-L-leucyl}amino)-1-hydroxy-3-[(3S)-2-oxopyrrolidin-3-yl]propane-1-sulfonic acid, (1S,2S)-2-({N-[(benzyloxy)carbonyl]-L-leucyl}amino)-1-hydroxy-3-[(3S)-2-oxopyrrolidin-3-yl]propane-1-sulfonic acid, 3C-like proteinase, ... | | Authors: | Wang, Y.C, Yang, C.S, Hou, M.H, Tsai, C.L, Chou, Y.Z, Chen, Y, Hung, M.C. | | Deposit date: | 2020-06-10 | | Release date: | 2021-05-05 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.69 Å) | | Cite: | Structural basis of SARS-CoV-2 main protease inhibition by a broad-spectrum anti-coronaviral drug.
Am J Cancer Res, 10, 2020
|
|
3T1I
 
 | | Crystal Structure of Human Mre11: Understanding Tumorigenic Mutations | | Descriptor: | 2,3-DIHYDROXY-1,4-DITHIOBUTANE, Double-strand break repair protein MRE11A, GLYCEROL, ... | | Authors: | Park, Y.B, Chae, J, Kim, Y, Cho, Y. | | Deposit date: | 2011-07-22 | | Release date: | 2011-11-30 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Crystal structure of human mre11: understanding tumorigenic mutations
Structure, 19, 2011
|
|
6XJP
 
 | |
4OO6
 
 | | Crystal structure of human KAP-beta2 bound to the NLS of HCC1 (Hepato Cellular Carcinoma protein 1) | | Descriptor: | RNA-binding protein 39, Transportin-1 | | Authors: | Sampathkumar, P, Brower, A, Soniat, M, Bonanno, J, Hillerich, B, Seidel, R.D, Rout, M.P, Chook, Y.M, Almo, S.C, New York Structural Genomics Research Consortium (NYSGRC), Nucleocytoplasmic Transport: a Target for Cellular Control (NPCXstals) | | Deposit date: | 2014-01-30 | | Release date: | 2014-02-12 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Crystal structure of human KAP-beta2 bound to the NLS of HCC1 (Hepato Cellular Carcinoma protein 1)
to be published
|
|
3TAI
 
 | | Crystal structure of NurA | | Descriptor: | DNA double-strand break repair protein nurA, GLYCEROL | | Authors: | Chae, J, Kim, Y.C, Cho, Y. | | Deposit date: | 2011-08-04 | | Release date: | 2011-11-23 | | Last modified: | 2013-07-03 | | Method: | X-RAY DIFFRACTION (2.82 Å) | | Cite: | Crystal structure of the NurA-dAMP-Mn2+ complex
Nucleic Acids Res., 40, 2012
|
|
3TAL
 
 | | Crystal structure of NurA with manganese | | Descriptor: | DNA double-strand break repair protein nurA, GLYCEROL, MANGANESE (II) ION | | Authors: | Chae, J, Kim, Y.C, Cho, Y. | | Deposit date: | 2011-08-04 | | Release date: | 2011-11-23 | | Last modified: | 2013-07-03 | | Method: | X-RAY DIFFRACTION (3.15 Å) | | Cite: | Crystal structure of the NurA-dAMP-Mn2+ complex
Nucleic Acids Res., 40, 2012
|
|
3TAZ
 
 | | Crystal structure of NurA bound to dAMP and manganese | | Descriptor: | 2'-DEOXYADENOSINE-5'-MONOPHOSPHATE, DNA double-strand break repair protein nurA, GLYCEROL, ... | | Authors: | Chae, J, Kim, Y.C, Cho, Y. | | Deposit date: | 2011-08-04 | | Release date: | 2011-11-23 | | Last modified: | 2024-06-05 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Crystal structure of the NurA-dAMP-Mn2+ complex
Nucleic Acids Res., 40, 2012
|
|
4YVM
 
 | |
8DYO
 
 | | Cryo-EM structure of Importin-4 bound to RanGTP | | Descriptor: | GTP-binding nuclear protein GSP1/CNR1, GUANOSINE-5'-TRIPHOSPHATE, Importin-4, ... | | Authors: | Bernardes, N.E, Fung, H.Y.J, Li, Y, Chen, Z, Chook, Y.M. | | Deposit date: | 2022-08-04 | | Release date: | 2022-09-21 | | Last modified: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (7.1 Å) | | Cite: | Structure of IMPORTIN-4 bound to the H3-H4-ASF1 histone-histone chaperone complex.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
4WVF
 
 | | Crystal structure of KPT276 in complex with CRM1-Ran-RanBP1 | | Descriptor: | (2E)-3-{3-[3,5-bis(trifluoromethyl)phenyl]-1H-1,2,4-triazol-1-yl}-1-(3,3-difluoroazetidin-1-yl)prop-2-en-1-one, 1,2-ETHANEDIOL, CHLORIDE ION, ... | | Authors: | Sun, Q, Chook, Y. | | Deposit date: | 2014-11-05 | | Release date: | 2015-07-15 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Nuclear export inhibitors avert progression in preclinical models of inflammatory demyelination.
Nat.Neurosci., 18, 2015
|
|
5JLJ
 
 | | Crystal Structure of KPT8602 in complex with CRM1-Ran-RanBP1 | | Descriptor: | (2R)-3-{3-[3,5-bis(trifluoromethyl)phenyl]-1H-1,2,4-triazol-1-yl}-2-(pyrimidin-5-yl)propanamide, CHLORIDE ION, Exportin-1, ... | | Authors: | Fung, H.Y, Chook, Y.M. | | Deposit date: | 2016-04-27 | | Release date: | 2016-06-29 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Next-generation XPO1 inhibitor shows improved efficacy and in vivo tolerability in hematological malignancies.
Leukemia, 30, 2016
|
|
5YVG
 
 | |
5YVH
 
 | |
5YVI
 
 | |
5J3V
 
 | |
6CIT
 
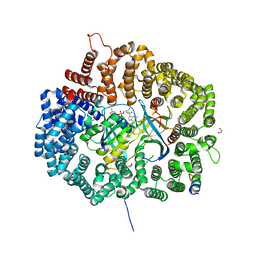 | |
6XJR
 
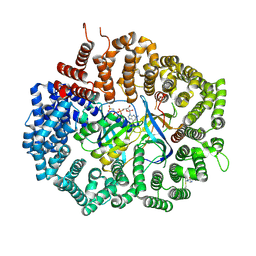 | |
6XJU
 
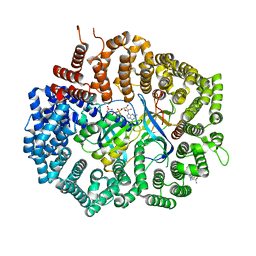 | | Crystal Structure of KPT-8602 bound to CRM1 (E582K, 537-DLTVK-541 to GLCEQ) | | Descriptor: | (2R)-3-{3-[3,5-bis(trifluoromethyl)phenyl]-1H-1,2,4-triazol-1-yl}-2-(pyrimidin-5-yl)propanamide, Exportin-1, GTP-binding nuclear protein Ran, ... | | Authors: | Baumhardt, J.M, Chook, Y.M. | | Deposit date: | 2020-06-24 | | Release date: | 2021-01-27 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.193 Å) | | Cite: | Recurrent XPO1 mutations alter pathogenesis of chronic lymphocytic leukemia.
J Hematol Oncol, 14, 2021
|
|
5DHA
 
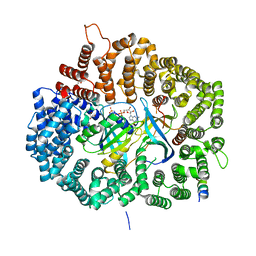 | |
5DIF
 
 | |
5DI9
 
 | |
6XJS
 
 | |
6XJT
 
 | | Crystal Structure of KPT-8602 bound to CRM1 (537-DLTVK-541 to GLCEQ) | | Descriptor: | (2R)-3-{3-[3,5-bis(trifluoromethyl)phenyl]-1H-1,2,4-triazol-1-yl}-2-(pyrimidin-5-yl)propanamide, Exportin-1, GTP-binding nuclear protein Ran, ... | | Authors: | Baumhardt, J.M, Chook, Y.M. | | Deposit date: | 2020-06-24 | | Release date: | 2021-01-27 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.406 Å) | | Cite: | Recurrent XPO1 mutations alter pathogenesis of chronic lymphocytic leukemia.
J Hematol Oncol, 14, 2021
|
|
