4JGJ
 
 | | Crystal structure of the Ig-like D1 domain from mouse Carcinoembryogenic antigen-related cell adhesion molecule 15 (CEACAM15) [PSI-NYSGRC-005691] | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Carcinoembryonic antigen-related cell adhesion molecule 15, Unknown peptide | | Authors: | Kumar, P.R, Bonanno, J, Ahmed, M, Banu, R, Bhosle, R, Calarese, D, Celikigil, A, Chamala, S, Chan, M.K, Chowdhury, S, Fiser, A, Garforth, S, Glenn, A.S, Hillerich, B, Khafizov, K, Love, J, Patel, H, Seidel, R, Stead, M, Toro, R, Nathenson, S.G, Almo, S.C, New York Structural Genomics Research Consortium (NYSGRC), Atoms-to-Animals: The Immune Function Network (IFN) | | Deposit date: | 2013-03-01 | | Release date: | 2013-03-13 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.6508 Å) | | Cite: | Crystal structure of the Ig-like D1 domain of CEACAM15 from Mus musculus [NYSGRC-005691]
to be published
|
|
5XEP
 
 | | Crystal structure of BRP39, a chitinase-like protein, at 2.6 Angstorm resolution | | Descriptor: | 1,2-ETHANEDIOL, Chitinase-3-like protein 1 | | Authors: | Mohanty, A.K, Fisher, A.J, Choudhary, S, Kaushik, J.K. | | Deposit date: | 2017-04-05 | | Release date: | 2018-04-11 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Crystal Structure of BRP39, a signalling glycoprotein expressed during mammary gland apoptosis.
To be published
|
|
1CNU
 
 | |
5LAO
 
 | | S-nitrosylated 3D NMR structure of the cytoplasmic rhodanese domain of the inner membrane protein YgaP from Escherichia coli | | Descriptor: | Inner membrane protein YgaP | | Authors: | Eichmann, C, Tzitzilonis, C, Nakamura, T, Maslennikov, I, Kwiatkowski, W, Choe, S, Lipton, S.A, Guntert, P, Riek, R. | | Deposit date: | 2016-06-14 | | Release date: | 2016-08-17 | | Last modified: | 2024-06-19 | | Method: | SOLUTION NMR | | Cite: | S-Nitrosylation Induces Structural and Dynamical Changes in a Rhodanese Family Protein.
J.Mol.Biol., 428, 2016
|
|
8FF2
 
 | |
5LAM
 
 | | Refined 3D NMR structure of the cytoplasmic rhodanese domain of the inner membrane protein YgaP from Escherichia coli | | Descriptor: | Inner membrane protein YgaP | | Authors: | Eichmann, C, Tzitzilonis, C, Nakamura, T, Maslennikov, I, Kwiatkowski, W, Choe, S, Lipton, S.A, Guntert, P, Riek, R. | | Deposit date: | 2016-06-14 | | Release date: | 2016-08-17 | | Last modified: | 2024-06-19 | | Method: | SOLUTION NMR | | Cite: | S-Nitrosylation Induces Structural and Dynamical Changes in a Rhodanese Family Protein.
J.Mol.Biol., 428, 2016
|
|
1YGM
 
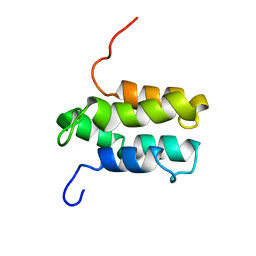 | | NMR structure of Mistic | | Descriptor: | hypothetical protein BSU31320 | | Authors: | Roosild, T.P, Greenwald, J, Vega, M, Castronovo, S, Riek, R, Choe, S. | | Deposit date: | 2005-01-05 | | Release date: | 2005-03-01 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | NMR structure of Mistic, a membrane-integrating protein for membrane protein expression.
Science, 307, 2005
|
|
4KNP
 
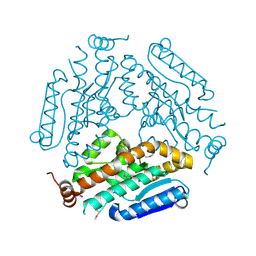 | | Crystal Structure Of a Putative enoyl-coA hydratase (PSI-NYSGRC-019597) from Mycobacterium avium paratuberculosis K-10 | | Descriptor: | enoyl-CoA hydratase | | Authors: | Kumar, P.R, Ahmed, M, Attonito, J, Bhosle, R, Chamala, S, Chowdhury, S, Glenn, A.S, Hammonds, J, Hillerich, B, Love, J.D, Seidel, R, Stead, M, Toro, R, Wasserman, S.R, Almo, S.C, New York Structural Genomics Research Consortium (NYSGRC) | | Deposit date: | 2013-05-10 | | Release date: | 2013-05-22 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (1.898 Å) | | Cite: | Crystal structure of a Putative enoyl-coA hydratase from Mycobacterium avium paratuberculosis K-10
to be published
|
|
8CU6
 
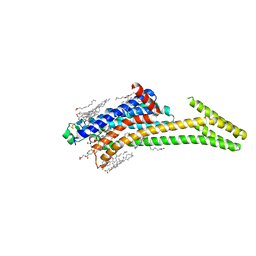 | | Crystal structure of A2AAR-StaR2-S277-bRIL in complex with a novel A2a antagonist, LJ-4517 | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, (2R,3R,4R)-2-[(8P)-6-amino-2-(hex-1-yn-1-yl)-8-(thiophen-2-yl)-9H-purin-9-yl]oxolane-3,4-diol, Adenosine receptor A2a,Soluble cytochrome b562, ... | | Authors: | Shiriaeva, A, Park, D.-J, Kim, G, Lee, Y, Hou, X, Jarhad, D.B, Kim, G, Yu, J, Hyun, Y.E, Kim, W, Gao, Z.-G, Jacobson, K.A, Han, G.W, Stevens, R.C, Jeong, L.S, Choi, S, Cherezov, V. | | Deposit date: | 2022-05-16 | | Release date: | 2022-08-31 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | GPCR Agonist-to-Antagonist Conversion: Enabling the Design of Nucleoside Functional Switches for the A 2A Adenosine Receptor.
J.Med.Chem., 65, 2022
|
|
8CU7
 
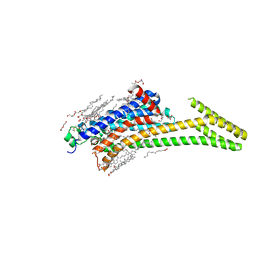 | | Crystal structure of A2AAR-StaR2-bRIL in complex with a novel A2a antagonist, LJ-4517 | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, (2R,3R,4R)-2-[(8P)-6-amino-2-(hex-1-yn-1-yl)-8-(thiophen-2-yl)-9H-purin-9-yl]oxolane-3,4-diol, Adenosine receptor A2a,Soluble cytochrome b562, ... | | Authors: | Shiriaeva, A, Park, D.-J, Kim, G, Lee, Y, Hou, X, Jarhad, D.B, Kim, G, Yu, J, Hyun, Y.E, Kim, W, Gao, Z.-G, Jacobson, K.A, Han, G.W, Stevens, R.C, Jeong, L.S, Choi, S, Cherezov, V. | | Deposit date: | 2022-05-16 | | Release date: | 2022-08-31 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | GPCR Agonist-to-Antagonist Conversion: Enabling the Design of Nucleoside Functional Switches for the A 2A Adenosine Receptor.
J.Med.Chem., 65, 2022
|
|
4KKN
 
 | | Crystal structure of bovine CTLA-4, PSI-NYSGRC-012704 | | Descriptor: | Cytotoxic T-lymphocyte associated protein 4, alpha-D-mannopyranose-(1-3)-beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose | | Authors: | Kumar, P.R, Ahmed, M, Banu, R, Bhosle, R, Bonanno, J, Calarese, D.A, Celikgil, A, Chamala, S, Chan, M.K, Chowdhury, S, Fiser, A, Garforth, S.J, Scott Glenn, A, Hammonds, J, Hillerich, B, Khafizov, K, Lafleur, J, Attonito, J, Love, J.D, Patel, H, Patel, R, Seidel, R.D, Smith, B, Stead, M, Toro, R, Casadevall, A, Almo, S.C, New York Structural Genomics Research Consortium (NYSGRC), Atoms-to-Animals: The Immune Function Network (IFN) | | Deposit date: | 2013-05-06 | | Release date: | 2013-06-12 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.253 Å) | | Cite: | Crystal structure of bovine CTLA-4, PSI-NYSGRC-012704
to be published
|
|
4KTO
 
 | | Crystal Structure Of a Putative Isovaleryl-CoA dehydrogenase (PSI-NYSGRC-012251) from Sinorhizobium meliloti 1021 | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, FLAVIN-ADENINE DINUCLEOTIDE, isovaleryl-CoA dehydrogenase | | Authors: | Kumar, P.R, Ahmed, M, Attonito, J, Bhosle, R, Bonanno, J, Chamala, S, Chowdhury, S, Glenn, A.S, Hammonds, J, Hillerich, B, Himmel, D, Love, J.D, Seidel, R, Stead, M, Toro, R, Wasserman, S.R, Almo, S.C, New York Structural Genomics Research Consortium (NYSGRC) | | Deposit date: | 2013-05-20 | | Release date: | 2013-06-19 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.137 Å) | | Cite: | Crystal structure of a Putative Isovaleryl-CoA dehydrogenase from Sinorhizobium meliloti 1021
to be published
|
|
3TVZ
 
 | | Structure of Bacillus subtilis HmoB | | Descriptor: | Putative uncharacterized protein yhgC | | Authors: | Choe, J, Choi, S, Park, S. | | Deposit date: | 2011-09-21 | | Release date: | 2012-07-11 | | Last modified: | 2019-10-09 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Bacillus subtilis HmoB is a heme oxygenase with a novel structure.
Bmb Rep, 45, 2012
|
|
1A68
 
 | |
1B8K
 
 | | Neurotrophin-3 from Human | | Descriptor: | PROTEIN (NEUROTROPHIN-3) | | Authors: | Robinson, R.C, Radziejewski, C, Stuart, D.I, Jones, E.Y, Choe, S. | | Deposit date: | 1999-02-01 | | Release date: | 1999-02-09 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | The structures of the neurotrophin 4 homodimer and the brain-derived neurotrophic factor/neurotrophin 4 heterodimer reveal a common Trk-binding site.
Protein Sci., 8, 1999
|
|
1B8M
 
 | | BRAIN DERIVED NEUROTROPHIC FACTOR, NEUROTROPHIN-4 | | Descriptor: | PROTEIN (BRAIN DERIVED NEUROTROPHIC FACTOR), PROTEIN (NEUROTROPHIN-4) | | Authors: | Robinson, R.C, Radziejewski, C, Stuart, D.I, Jones, E.Y, Choe, S. | | Deposit date: | 1999-02-01 | | Release date: | 1999-02-09 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | The structures of the neurotrophin 4 homodimer and the brain-derived neurotrophic factor/neurotrophin 4 heterodimer reveal a common Trk-binding site.
Protein Sci., 8, 1999
|
|
1B98
 
 | | NEUROTROPHIN 4 (HOMODIMER) | | Descriptor: | CHLORIDE ION, PROTEIN (NEUROTROPHIN-4) | | Authors: | Robinson, R.C, Radziejewski, C, Stuart, D.I, Jones, E.Y, Choe, S. | | Deposit date: | 1999-02-22 | | Release date: | 1999-02-26 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | The structures of the neurotrophin 4 homodimer and the brain-derived neurotrophic factor/neurotrophin 4 heterodimer reveal a common Trk-binding site.
Protein Sci., 8, 1999
|
|
1B9F
 
 | | MOBILITY OF AN HIV-1 INTEGRASE ACTIVE SITE LOOP IS CORRELATED WITH CATALYTIC ACTIVITY | | Descriptor: | CACODYLATE ION, PROTEIN (INTEGRASE), SULFATE ION | | Authors: | Greenwald, J, Le, V, Butler, S.L, Bushman, F.D, Choe, S. | | Deposit date: | 1999-02-11 | | Release date: | 1999-07-19 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | The mobility of an HIV-1 integrase active site loop is correlated with catalytic activity.
Biochemistry, 38, 1999
|
|
1B92
 
 | | MOBILITY OF AN HIV-1 INTEGRASE ACTIVE SITE LOOP IS CORRELATED WITH CATALYTIC ACTIVITY | | Descriptor: | CACODYLATE ION, PROTEIN (INTEGRASE), SULFATE ION | | Authors: | Greenwald, J, Le, V, Butler, S.L, Bushman, F.D, Choe, S. | | Deposit date: | 1999-02-19 | | Release date: | 1999-07-19 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.02 Å) | | Cite: | The mobility of an HIV-1 integrase active site loop is correlated with catalytic activity.
Biochemistry, 38, 1999
|
|
1B9D
 
 | | MOBILITY OF AN HIV-1 INTEGRASE ACTIVE SITE LOOP IS CORRELATED WITH CATALYTIC ACTIVITY | | Descriptor: | CACODYLATE ION, PROTEIN (INTEGRASE), SULFATE ION | | Authors: | Greenwald, J, Le, V, Butler, S.L, Bushman, F.D, Choe, S. | | Deposit date: | 1999-02-11 | | Release date: | 1999-07-19 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | The mobility of an HIV-1 integrase active site loop is correlated with catalytic activity.
Biochemistry, 38, 1999
|
|
5Z81
 
 | |
1BTE
 
 | | CRYSTAL STRUCTURE OF THE EXTRACELLULAR DOMAIN OF THE TYPE II ACTIVIN RECEPTOR | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, PROTEIN (ACTIVIN RECEPTOR TYPE II) | | Authors: | Greenwald, J, Fischer, W, Vale, W, Choe, S. | | Deposit date: | 1998-09-01 | | Release date: | 1999-02-09 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Three-finger toxin fold for the extracellular ligand-binding domain of the type II activin receptor serine kinase.
Nat.Struct.Biol., 6, 1999
|
|
7TVH
 
 | |
6W18
 
 | | Structure of S. pombe Arp2/3 complex in inactive state | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, Actin-related protein 2, Actin-related protein 2/3 complex subunit 1, ... | | Authors: | Shaaban, M, Nolen, B.J, Chowdhury, S. | | Deposit date: | 2020-03-03 | | Release date: | 2020-08-12 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (4.2 Å) | | Cite: | Cryo-EM reveals the transition of Arp2/3 complex from inactive to nucleation-competent state.
Nat.Struct.Mol.Biol., 27, 2020
|
|
1T24
 
 | | Plasmodium falciparum lactate dehydrogenase complexed with NAD+ and 4-hydroxy-1,2,5-oxadiazole-3-carboxylic acid | | Descriptor: | 4-HYDROXY-1,2,5-OXADIAZOLE-3-CARBOXYLIC ACID, GLYCEROL, L-lactate dehydrogenase, ... | | Authors: | Cameron, A, Read, J, Tranter, R, Winter, V.J, Sessions, R.B, Brady, R.L, Vivas, L, Easton, A, Kendrick, H, Croft, S.L, Barros, D, Lavandera, J.L, Martin, J.J, Risco, F, Garcia-Ochoa, S, Gamo, F.J, Sanz, L, Leon, L, Ruiz, J.R, Gabarro, R, Mallo, A, De Las Heras, F.G. | | Deposit date: | 2004-04-20 | | Release date: | 2004-05-11 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Identification and Activity of a Series of Azole-based Compounds with Lactate Dehydrogenase-directed Anti-malarial Activity.
J.Biol.Chem., 279, 2004
|
|
